

You can:
| Name | 5-hydroxytryptamine receptor 2C |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR2C |
| Synonym | Serotonin receptor 2C serotonin 1c receptor 5-HT1C 5-HT2C 5-HT-2C [ Show all ] |
| Disease | Pain Sleep initiation and maintenance disorders; Primary insomnia; Schizophrenia Unspecified Depression Drug abuse [ Show all ] |
| Length | 458 |
| Amino acid sequence | MVNLRNAVHSFLVHLIGLLVWQCDISVSPVAAIVTDIFNTSDGGRFKFPDGVQNWPALSIVIIIIMTIGGNILVIMAVSMEKKLHNATNYFLMSLAIADMLVGLLVMPLSLLAILYDYVWPLPRYLCPVWISLDVLFSTASIMHLCAISLDRYVAIRNPIEHSRFNSRTKAIMKIAIVWAISIGVSVPIPVIGLRDEEKVFVNNTTCVLNDPNFVLIGSFVAFFIPLTIMVITYCLTIYVLRRQALMLLHGHTEEPPGLSLDFLKCCKRNTAEEENSANPNQDQNARRRKKKERRPRGTMQAINNERKASKVLGIVFFVFLIMWCPFFITNILSVLCEKSCNQKLMEKLLNVFVWIGYVCSGINPLVYTLFNKIYRRAFSNYLRCNYKVEKKPPVRQIPRVAATALSGRELNVNIYRHTNEPVIEKASDNEPGIEMQVENLELPVNPSSVVSERISSV |
| UniProt | P28335 |
| Protein Data Bank | 6bqg, 6bqh |
| GPCR-HGmod model | P28335 |
| 3D structure model | This structure is from PDB ID 6bqg. |
| BioLiP | BL0404805, BL0404806 |
| Therapeutic Target Database | T83813 |
| ChEMBL | CHEMBL225 |
| IUPHAR | 8 |
| DrugBank | BE0004957, BE0004881, BE0000533 |
| Name | 1-(3-Chlorophenyl)piperazine |
|---|---|
| Molecular formula | C10H13ClN2 |
| IUPAC name | 1-(3-chlorophenyl)piperazine |
| Molecular weight | 196.678 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 1 |
| XlogP | 1.7 |
| Synonyms | Piperazine, 1-(3-chlorophenyl)- AJ-08201 ST2403154 BBL008938 UNII-REY0CNO998 [ Show all ] |
| Inchi Key | VHFVKMTVMIZMIK-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C10H13ClN2/c11-9-2-1-3-10(8-9)13-6-4-12-5-7-13/h1-3,8,12H,4-7H2 |
| PubChem CID | 1355 |
| ChEMBL | CHEMBL478 |
| IUPHAR | 142 |
| BindingDB | 50001915 |
| DrugBank | DB12110 |
Structure | 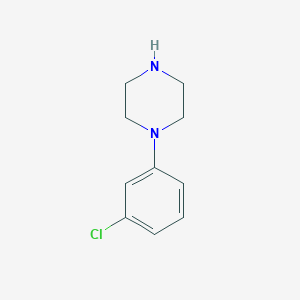 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| EC50 | 7.943 nM | PMID18095642 | ChEMBL |
| EC50 | 8.1 nM | PMID23537943 | ChEMBL |
| EC50 | 12.59 nM | PMID27864071 | ChEMBL |
| EC50 | 13.0 nM | PMID27864071 | BindingDB |
| EC50 | 15.0 nM | PMID20022752, PMID17315987 | BindingDB,ChEMBL |
| EC50 | 26.0 nM | PMID16257207, PMID15081042 | BindingDB,ChEMBL |
| EC50 | 170.0 nM | PMID19646865, PMID19716297 | BindingDB,ChEMBL |
| Efficacy | 83.0 % | PMID16257207 | ChEMBL |
| Emax | 71.0 % | PMID19646865, PMID19716297 | ChEMBL |
| Emax | 90.0 % | PMID27864071 | ChEMBL |
| Intrinsic activity | 1.0 - | PMID20022752 | ChEMBL |
| Ki | 3.16228 - 316.228 nM | PMID15322733, PMID9933142, PMID12970106, PMID10217294, PMID10611640, PMID14709324 | IUPHAR |
| Ki | 7.9 nM | PMID27864071 | BindingDB |
| Ki | 7.943 nM | PMID27864071 | ChEMBL |
| Ki | 9.0 nM | PMID16257207, PMID15081042 | PDSP,BindingDB,ChEMBL |
| Ki | 14.1254 nM | PMID15322733 | PDSP |
| Ki | 14.13 nM | PMID15322733 | BindingDB |
| Ki | 16.0 nM | PMID14709324 | PDSP,BindingDB |
| Ki | 17.0 nM | PMID20022752, PMID17315987 | BindingDB,ChEMBL |
| Ki | 27.7 nM | PMID26748694 | ChEMBL |
| Ki | 28.0 nM | PMID26748694 | BindingDB |
| Ki | 81.28 nM | PMID10498829 | BindingDB |
| Ki | 81.2831 nM | PMID10498829 | PDSP |
| Ki | 85.11 nM | PMID11882920 | BindingDB |
| Ki | 85.1138 nM | PMID11882920 | PDSP |
| Ki | 104.71 nM | PMID9225287 | PDSP,BindingDB |
| Ki | 251.18 nM | PMID7582481 | PDSP,BindingDB |
| Ki | 360.0 nM | PMID2537663 | BindingDB |
| Relative efficacy | 83.0 % | PMID15081042 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417