

You can:
| Name | Adenosine receptor A1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Adora1 |
| Synonym | A1 receptor A1-AR A1R adenosine receptor A1 RDC7 |
| Disease | N/A for non-human GPCRs |
| Length | 326 |
| Amino acid sequence | MPPYISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYFHTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKTVVTQRRAAVAIAGCWILSLVVGLTPMFGWNNLSVVEQDWRANGSVGEPVIKCEFEKVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLNKKVSASSGDPQKYYGKELKIAKSLALILFLFALSWLPLHILNCITLFCPTCQKPSILIYIAIFLTHGNSAMNPIVYAFRIHKFRVTFLKIWNDHFRCQPKPPIDEDLPEEKAED |
| UniProt | P25099 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL318 |
| IUPHAR | 18 |
| DrugBank | N/A |
| Name | N6-Cyclopentyladenosine |
|---|---|
| Molecular formula | C15H21N5O4 |
| IUPAC name | (2R,3R,4S,5R)-2-[6-(cyclopentylamino)purin-9-yl]-5-(hydroxymethyl)oxolane-3,4-diol |
| Molecular weight | 335.364 |
| Hydrogen bond acceptor | 8 |
| Hydrogen bond donor | 4 |
| XlogP | 0.9 |
| Synonyms | SQMWSBKSHWARHU-SDBHATRESA-N 5720AB J-523544 N(6)-Cyclopentyladenosine NCGC00023909-04 [ Show all ] |
| Inchi Key | SQMWSBKSHWARHU-SDBHATRESA-N |
| Inchi ID | InChI=1S/C15H21N5O4/c21-5-9-11(22)12(23)15(24-9)20-7-18-10-13(16-6-17-14(10)20)19-8-3-1-2-4-8/h6-9,11-12,15,21-23H,1-5H2,(H,16,17,19)/t9-,11-,12-,15-/m1/s1 |
| PubChem CID | 657378 |
| ChEMBL | CHEMBL68738 |
| IUPHAR | 380 |
| BindingDB | 25400 |
| DrugBank | N/A |
Structure | 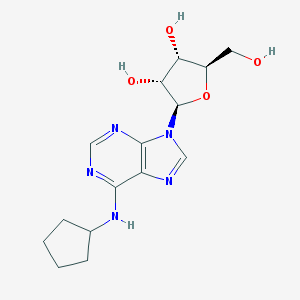 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 86.18 % | PMID17689079 | ChEMBL |
| Activity | 100.0 % | PMID9438026 | ChEMBL |
| Activity | 100.0 - | PMID11520205 | ChEMBL |
| Activity | 193.0 % | PMID12398547 | ChEMBL |
| EC25 | 3.0 nM | PMID3336027 | ChEMBL |
| EC50 | 13.3 nM | PMID10649980 | BindingDB,ChEMBL |
| EC50 | 14.0 nM | PMID9438026 | BindingDB |
| EC50 | 14.3 nM | PMID9438026 | ChEMBL |
| EC50 | 16.9 nM | PMID12398547 | BindingDB,ChEMBL |
| EC50 | 20.0 nM | PMID10841798 | BindingDB |
| EC50 | 20.3 nM | PMID10841798 | ChEMBL |
| GTP shift | 6.0 - | PMID8691477 | ChEMBL |
| IC50 | 0.32 nM | PMID1542091 | BindingDB |
| IC50 | 0.3236 nM | PMID1542091 | ChEMBL |
| IC50 | 0.35 nM | PMID3351850 | BindingDB,ChEMBL |
| IC50 | 1.0 nM | PMID1875349 | BindingDB,ChEMBL |
| IC50 | 7.09 nM | PMID20188574 | ChEMBL |
| IC50 | 7.1 nM | PMID20188574 | BindingDB |
| IC50 | 58.0 nM | PMID3373486 | BindingDB,ChEMBL |
| IC50 | 154.0 nM | PMID20188574 | BindingDB,ChEMBL |
| IC50 | 640.0 nM | PMID2374150 | BindingDB,ChEMBL |
| IC50 | 720.0 nM | PMID3351850 | BindingDB,ChEMBL |
| IC50 | 11000.0 nM | PMID3351851 | BindingDB,ChEMBL |
| IC50 | 14000.0 nM | PMID3351851 | BindingDB,ChEMBL |
| Inhibition | 0.99 % | PMID3373486 | ChEMBL |
| Ki | 0.3 nM | PMID11881988 | BindingDB,ChEMBL |
| Ki | 0.32 nM | PMID1554381, PMID2258897 | BindingDB,ChEMBL |
| Ki | 0.45 nM | PMID1766003 | BindingDB,ChEMBL |
| Ki | 0.589 nM | PMID2374150 | ChEMBL |
| Ki | 0.59 nM | PMID3336027, PMID1738138, PMID2909748, PMID2995663, PMID3385722, PMID7932588, PMID1548682, PMID1433217, PMID2754691, PMID2374150 | BindingDB,ChEMBL |
| Ki | 0.59 nM | PMID3336027, PMID3385722, PMID2995663 | BindingDB |
| Ki | 0.6 nM | Bioorg. Med. Chem. Lett., (1991) 1:9:481 | ChEMBL |
| Ki | 0.6 nM | N/A | BindingDB |
| Ki | 0.76 nM | PMID1495019 | BindingDB |
| Ki | 0.8 nM | PMID20188574, PMID1619615, PMID3373486 | BindingDB |
| Ki | 0.8 nM | PMID20188574, PMID3216901, PMID1619615, PMID3373486 | BindingDB,ChEMBL |
| Ki | 0.8 nM | PMID1619615 | BindingDB |
| Ki | 1.2 nM | , PMID1732541, PMID10479279, Bioorg. Med. Chem. Lett., (1993) 3:12:2661 | BindingDB,ChEMBL |
| Ki | 1.21 nM | PMID1732541 | ChEMBL |
| Ki | 1.5 nM | PMID10841798 | BindingDB,ChEMBL |
| Ki | 4.4 nM | PMID26392370 | BindingDB |
| Ki | 4.43 nM | PMID26392370 | ChEMBL |
| Ki | 5.07 nM | PMID10649980 | BindingDB,ChEMBL |
| Ki | 5.9 nM | PMID7562934, PMID10212125, PMID26392370, PMID9438026, PMID8691477, PMID10999489 | BindingDB,ChEMBL |
| Ki | 7.14 nM | PMID11520205 | BindingDB,ChEMBL |
| Ki | 7.9 nM | PMID26392370 | BindingDB,ChEMBL |
| Ki | 8.48 nM | PMID26776359 | ChEMBL |
| Ki | 8.5 nM | PMID26776359 | BindingDB |
| Ki | 10.4 nM | PMID26462195 | ChEMBL |
| Ki | 15.0 nM | PMID26392370 | BindingDB |
| Ki | 15.3 nM | PMID26392370 | ChEMBL |
| Ki | 35.0 nM | PMID26392370, PMID9438026, PMID7562934, PMID8691477 | BindingDB,ChEMBL |
| Ki | 35.2 nM | PMID26392370, PMID9438026, PMID7562934 | ChEMBL |
| Ki | 63.0 nM | PMID26392370 | BindingDB |
| Ki | 63.08 nM | PMID26392370 | ChEMBL |
| Ki | 99.0 nM | PMID26392370 | BindingDB |
| Ki | 99.21 nM | PMID26392370 | ChEMBL |
| Ki | 130.0 nM | PMID1619615 | BindingDB,ChEMBL |
| Ki | 230.0 nM | PMID11277527 | BindingDB,ChEMBL |
| Ki | 462.0 nM | PMID2374150 | BindingDB,ChEMBL |
| Max | 100.0 % | PMID10841798 | ChEMBL |
| Ratio IC50 | 21.9 - | PMID20188574 | ChEMBL |
| Ratio Ki | 6.0 - | PMID26392370 | ChEMBL |
| Ratio Ki | 6.48 - | PMID26392370 | ChEMBL |
| Ratio Ki | 14.24 - | PMID26392370 | ChEMBL |
| RIA | 101.6 % | PMID10649980 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417