

You can:
| Name | Muscarinic acetylcholine receptor M1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Chrm1 |
| Synonym | cholinergic receptor, muscarinic 1 cholinergic receptor, muscarinic 1, CNS cholinergic receptor M1 muscarinic acetylcholine receptor M1 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 460 |
| Amino acid sequence | MNTSVPPAVSPNITVLAPGKGPWQVAFIGITTGLLSLATVTGNLLVLISFKVNTELKTVNNYFLLSLACADLIIGTFSMNLYTTYLLMGHWALGTLACDLWLALDYVASNASVMNLLLISFDRYFSVTRPLSYRAKRTPRRAALMIGLAWLVSFVLWAPAILFWQYLVGERTVLAGQCYIQFLSQPIITFGTAMAAFYLPVTVMCTLYWRIYRETENRARELAALQGSETPGKGGGSSSSSERSQPGAEGSPESPPGRCCRCCRAPRLLQAYSWKEEEEEDEGSMESLTSSEGEEPGSEVVIKMPMVDSEAQAPTKQPPKSSPNTVKRPTKKGRDRGGKGQKPRGKEQLAKRKTFSLVKEKKAARTLSAILLAFILTWTPYNIMVLVSTFCKDCVPETLWELGYWLCYVNSTVNPMCYALCNKAFRDTFRLLLLCRWDKRRWRKIPKRPGSVHRTPSRQC |
| UniProt | P08482 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL276 |
| IUPHAR | 13 |
| DrugBank | N/A |
| Name | Oxotremorine |
|---|---|
| Molecular formula | C12H18N2O |
| IUPAC name | 1-(4-pyrrolidin-1-ylbut-2-ynyl)pyrrolidin-2-one |
| Molecular weight | 206.289 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 0 |
| XlogP | 0.4 |
| Synonyms | KBio2_002393 KBioSS_002398 NCGC00015773-03 NSC-330497 SMR000528860 [ Show all ] |
| Inchi Key | RSDOPYMFZBJHRL-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C12H18N2O/c15-12-6-5-11-14(12)10-4-3-9-13-7-1-2-8-13/h1-2,5-11H2 |
| PubChem CID | 4630 |
| ChEMBL | CHEMBL7634 |
| IUPHAR | 302 |
| BindingDB | 50004665 |
| DrugBank | N/A |
Structure | 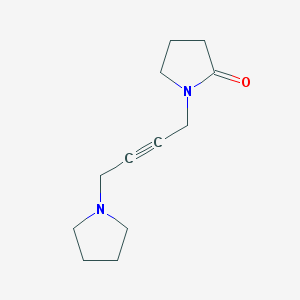 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 100.0 nM | , Bioorg. Med. Chem. Lett., (1997) 7:8:1033 | BindingDB,ChEMBL |
| IC50 | 113.0 nM | PMID1732522 | BindingDB,ChEMBL |
| IC50 | 360.0 nM | , Bioorg. Med. Chem. Lett., (1991) 1:3:147 | BindingDB,ChEMBL |
| Ki | 120.0 nM | PMID8246221 | BindingDB,ChEMBL |
| Ki | 196.0 nM | PMID1578480 | BindingDB,ChEMBL |
| Ki | 490.0 nM | PMID1310135 | BindingDB |
| Ki | 1000.0 nM | PMID2537406 | IUPHAR |
| Ki | 1.58489e+15 nM | PMID9703467 | ChEMBL |
| pD2 | 7.1 - | PMID1732522 | ChEMBL |
| pD2 | 8.0 - | PMID1732522 | ChEMBL |
| Ratio | 0.03 - | PMID2258905 | ChEMBL |
| Ratio | 1.0 - | PMID2258905 | ChEMBL |
| Ratio | 40.4 - | PMID1732522 | ChEMBL |
| Response | 13.7 % | PMID1578480 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417