

You can:
| Name | Muscarinic acetylcholine receptor M1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM1 |
| Synonym | cholinergic receptor cholinergic receptor, muscarinic 1 cholinergic receptor, muscarinic 1, CNS Chrm-1 M1 receptor [ Show all ] |
| Disease | Functional bowel syndrome; Irritable bowel syndrome Glaucoma Peptic ulcer Parkinsonism; Extrapyramidal disorders secondary to neuroleptic drug therapy Visceral spasms [ Show all ] |
| Length | 460 |
| Amino acid sequence | MNTSAPPAVSPNITVLAPGKGPWQVAFIGITTGLLSLATVTGNLLVLISFKVNTELKTVNNYFLLSLACADLIIGTFSMNLYTTYLLMGHWALGTLACDLWLALDYVASNASVMNLLLISFDRYFSVTRPLSYRAKRTPRRAALMIGLAWLVSFVLWAPAILFWQYLVGERTVLAGQCYIQFLSQPIITFGTAMAAFYLPVTVMCTLYWRIYRETENRARELAALQGSETPGKGGGSSSSSERSQPGAEGSPETPPGRCCRCCRAPRLLQAYSWKEEEEEDEGSMESLTSSEGEEPGSEVVIKMPMVDPEAQAPTKQPPRSSPNTVKRPTKKGRDRAGKGQKPRGKEQLAKRKTFSLVKEKKAARTLSAILLAFILTWTPYNIMVLVSTFCKDCVPETLWELGYWLCYVNSTINPMCYALCNKAFRDTFRLLLLCRWDKRRWRKIPKRPGSVHRTPSRQC |
| UniProt | P11229 |
| Protein Data Bank | 5cxv |
| GPCR-HGmod model | P11229 |
| 3D structure model | This structure is from PDB ID 5cxv. |
| BioLiP | BL0339262, BL0339261, BL0339263 |
| Therapeutic Target Database | T28893 |
| ChEMBL | CHEMBL216 |
| IUPHAR | 13 |
| DrugBank | BE0000092 |
| Name | pirenzepine |
|---|---|
| Molecular formula | C19H21N5O2 |
| IUPAC name | 11-[2-(4-methylpiperazin-1-yl)acetyl]-5H-pyrido[2,3-b][1,4]benzodiazepin-6-one |
| Molecular weight | 351.41 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 1 |
| XlogP | 0.1 |
| Synonyms | C19H21N5O2 Spectrum3_001453 CTK6I3129 UNII-3G0285N20N DSSTox_RID_77049 [ Show all ] |
| Inchi Key | RMHMFHUVIITRHF-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C19H21N5O2/c1-22-9-11-23(12-10-22)13-17(25)24-16-7-3-2-5-14(16)19(26)21-15-6-4-8-20-18(15)24/h2-8H,9-13H2,1H3,(H,21,26) |
| PubChem CID | 4848 |
| ChEMBL | CHEMBL9967 |
| IUPHAR | 328 |
| BindingDB | 39341 |
| DrugBank | DB00670 |
Structure | 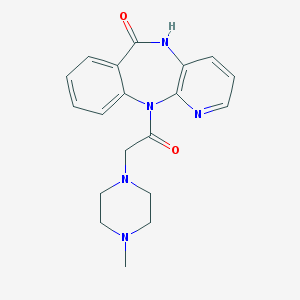 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| IC50 | 1.479 nM | PMID25275964 | ChEMBL |
| IC50 | 1.5 nM | PMID25275964 | BindingDB |
| IC50 | 2.1 nM | PMID26988801, PMID27876250 | BindingDB |
| IC50 | 12.0 nM | PMID23466604, PMID18588282 | ChEMBL |
| IC50 | 19.0 nM | PMID20875743 | BindingDB,ChEMBL |
| IC50 | 20.0 nM | PMID25275964 | BindingDB |
| IC50 | 20.42 nM | PMID25275964 | ChEMBL |
| IC50 | 21.0 nM | PMID26988801, PMID27876250 | ChEMBL |
| IC50 | 27.0 nM | PMID23582449 | ChEMBL |
| IC50 | 28.0 nM | PMID9986705 | BindingDB |
| IC50 | 28.2 nM | PMID9986705 | ChEMBL |
| IC50 | 29.0 nM | PMID18983139 | ChEMBL |
| IC50 | 48.98 nM | PMID25275964 | ChEMBL |
| IC50 | 49.0 nM | PMID25275964 | BindingDB |
| IC50 | 119.0 nM | PMID10522693 | BindingDB,ChEMBL |
| Imax | 83.0 % | PMID25275964 | ChEMBL |
| Imax | >100.0 % | PMID25275964 | ChEMBL |
| Inhibition | 96.7 % | PMID25765911 | ChEMBL |
| KA | 0.011 uM | PMID1379640 | ChEMBL |
| Kd | 20.0 nM | PMID15294002 | BindingDB,ChEMBL |
| Ki | 2.818 nM | PMID18595721 | ChEMBL |
| Ki | 2.82 nM | PMID18595721 | BindingDB |
| Ki | 4.6 nM | , Bioorg. Med. Chem. Lett., (1996) 6:7:785 | BindingDB,ChEMBL |
| Ki | 5.01187 - 15.8489 nM | PMID12049493, PMID2704370, PMID7925952, PMID1994002, PMID9113359, PMID2043926 | IUPHAR |
| Ki | 6.30957 nM | PMID1994002, PMID8016895 | PDSP |
| Ki | 6.31 nM | PMID1994002 | BindingDB |
| Ki | 8.0 nM | PMID1346637 | PDSP,BindingDB |
| Ki | 9.1 nM | PMID23379472 | BindingDB,ChEMBL |
| Ki | 10.0 nM | PMID23466604 | ChEMBL |
| Ki | 11.0 nM | PMID9121349 | PDSP |
| Ki | 13.0 nM | PMID1704434 | PDSP,BindingDB |
| Ki | 14.0 nM | PMID7853341 | BindingDB,ChEMBL |
| Ki | 15.0 nM | PMID25557493, PMID2385234, PMID24805037 | PDSP,BindingDB,ChEMBL |
| Ki | 16.0 nM | PMID25557493 | BindingDB,ChEMBL |
| Ki | 21.3796 nM | PMID13679167 | PDSP |
| Ki | 21.38 nM | PMID13679167, PMID17889543 | BindingDB,ChEMBL |
| Ki | 23.5 nM | PMID16302823, PMID15294002 | BindingDB,ChEMBL |
| Ki | 24.0 nM | PMID15294002 | BindingDB |
| Ki | 25.0 nM | PMID18983139 | ChEMBL |
| Ki | 33.0 nM | PMID2537406 | PDSP,BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417