

You can:
| Name | 5-hydroxytryptamine receptor 1A |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Htr1a |
| Synonym | 5-HT1A receptor 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled 5-HT1A ADRB2RL1 ADRBRL1 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 422 |
| Amino acid sequence | MDVFSFGQGNNTTASQEPFGTGGNVTSISDVTFSYQVITSLLLGTLIFCAVLGNACVVAAIALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPEDRSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVRKVEKKGAGTSLGTSSAPPPKKSLNGQPGSGDWRRCAENRAVGTPCTNGAVRQGDDEATLEVIEVHRVGNSKEHLPLPSESGSNSYAPACLERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHMPALLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFCRR |
| UniProt | P19327 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL273 |
| IUPHAR | 1 |
| DrugBank | N/A |
| Name | serotonin |
|---|---|
| Molecular formula | C10H12N2O |
| IUPAC name | 3-(2-aminoethyl)-1H-indol-5-ol |
| Molecular weight | 176.219 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 3 |
| XlogP | 0.2 |
| Synonyms | NCGC00142449-02 Bio2_000396 Prestwick1_000481 BSPBio_001112 SMP1_000272 [ Show all ] |
| Inchi Key | QZAYGJVTTNCVMB-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C10H12N2O/c11-4-3-7-6-12-10-2-1-8(13)5-9(7)10/h1-2,5-6,12-13H,3-4,11H2 |
| PubChem CID | 5202 |
| ChEMBL | CHEMBL39 |
| IUPHAR | 5 |
| BindingDB | 10755 |
| DrugBank | DB08839 |
Structure | 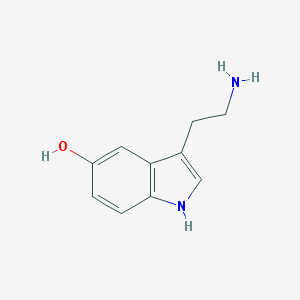 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 100.0 % | PMID18562201 | ChEMBL |
| EC50 | 10.0 nM | PMID8057297 | BindingDB,ChEMBL |
| EC50 | 323.0 nM | PMID18598015 | BindingDB,ChEMBL |
| Emax | 100.0 % | PMID18598015 | ChEMBL |
| IC50 | 2.6 nM | PMID9357534 | BindingDB,ChEMBL |
| IC50 | 3.0 nM | PMID9046348 | BindingDB,ChEMBL |
| IC50 | 5.2 nM | , PMID2374139, PMID8057297, Bioorg. Med. Chem. Lett., (1995) 5:20:2391 | BindingDB,ChEMBL |
| Ki | 0.12 nM | PMID9400006 | BindingDB |
| Ki | 1.585 nM | PMID10543880 | ChEMBL |
| Ki | 1.6 nM | PMID19425598, PMID19831400 | BindingDB,ChEMBL |
| Ki | 1.7 nM | PMID2965244 | BindingDB,ChEMBL |
| Ki | 1.862 nM | PMID17803293 | ChEMBL |
| Ki | 2.0 nM | PMID2299641 | BindingDB,ChEMBL |
| Ki | 2.1 nM | PMID23524160, PMID7629808 | BindingDB,ChEMBL |
| Ki | 2.6 nM | PMID21816515 | BindingDB,ChEMBL |
| Ki | 3.0 nM | PMID3543362, PMID18562201 | BindingDB,ChEMBL |
| Ki | 4.2 nM | PMID8584042 | BindingDB |
| Ki | 4.3 nM | PMID8230102 | BindingDB |
| Ki | 4.31 nM | PMID8230102 | ChEMBL |
| Ki | 4.786 nM | PMID8568799 | ChEMBL |
| Ki | 7.3 nM | PMID10229626, PMID9513601 | BindingDB,ChEMBL |
| Ki | 9.0 nM | Med Chem Res, (2008) 17:8:507 | ChEMBL |
| Ki | 46.77 nM | PMID6225026 | BindingDB |
| Ki | 90.0 nM | N/A | BindingDB |
| Ki | 134.0 nM | PMID8398139 | BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417