

You can:
| Name | 5-hydroxytryptamine receptor 2A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR2A |
| Synonym | 5-HT-2 serotonin receptor 2A serotonin 5HT-2 receptor 5-HT-2A 5-HT2A receptor [ Show all ] |
| Disease | Depression Unspecified Diabetes Erythropoietic porphyria Fibromyalgia [ Show all ] |
| Length | 471 |
| Amino acid sequence | MDILCEENTSLSSTTNSLMQLNDDTRLYSNDFNSGEANTSDAFNWTVDSENRTNLSCEGCLSPSCLSLLHLQEKNWSALLTAVVIILTIAGNILVIMAVSLEKKLQNATNYFLMSLAIADMLLGFLVMPVSMLTILYGYRWPLPSKLCAVWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNSRTKAFLKIIAVWTISVGISMPIPVFGLQDDSKVFKEGSCLLADDNFVLIGSFVSFFIPLTIMVITYFLTIKSLQKEATLCVSDLGTRAKLASFSFLPQSSLSSEKLFQRSIHREPGSYTGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICKESCNEDVIGALLNVFVWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENKKPLQLILVNTIPALAYKSSQLQMGQKKNSKQDAKTTDNDCSMVALGKQHSEEASKDNSDGVNEKVSCV |
| UniProt | P28223 |
| Protein Data Bank | 6a93, 6a94 |
| GPCR-HGmod model | P28223 |
| 3D structure model | This structure is from PDB ID 6a93. |
| BioLiP | BL0441025,BL0441028, BL0441031, BL0441030,BL0441033, BL0441029,BL0441032, BL0441026, BL0441024,BL0441027 |
| Therapeutic Target Database | T32060 |
| ChEMBL | CHEMBL224 |
| IUPHAR | 6 |
| DrugBank | BE0000451 |
| Name | serotonin |
|---|---|
| Molecular formula | C10H12N2O |
| IUPAC name | 3-(2-aminoethyl)-1H-indol-5-ol |
| Molecular weight | 176.219 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 3 |
| XlogP | 0.2 |
| Synonyms | NCGC00142449-02 Bio2_000396 Prestwick1_000481 BSPBio_001112 SMP1_000272 [ Show all ] |
| Inchi Key | QZAYGJVTTNCVMB-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C10H12N2O/c11-4-3-7-6-12-10-2-1-8(13)5-9(7)10/h1-2,5-6,12-13H,3-4,11H2 |
| PubChem CID | 5202 |
| ChEMBL | CHEMBL39 |
| IUPHAR | 5 |
| BindingDB | 10755 |
| DrugBank | DB08839 |
Structure | 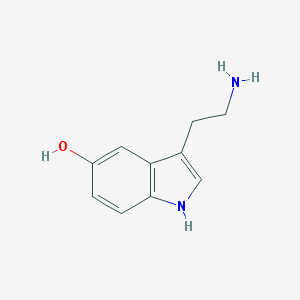 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Activity | <75.0 % | None | ChEMBL |
| Activity | 100.0 % | PMID27487565 | ChEMBL |
| Activity | 100.4 % | PMID23675993 | ChEMBL |
| EC50 | 1.5 nM | PMID23602445 | ChEMBL |
| EC50 | 1.549 nM | PMID23602445 | ChEMBL |
| EC50 | 1.88 nM | PMID25633969 | ChEMBL |
| EC50 | 1.9 nM | PMID25633969 | BindingDB |
| EC50 | 2.57 nM | PMID23301527 | ChEMBL |
| EC50 | 2.6 nM | PMID23301527 | BindingDB,ChEMBL |
| EC50 | 5.8 nM | PMID19284718 | BindingDB,ChEMBL |
| EC50 | 7.6 nM | PMID22778800 | BindingDB,ChEMBL |
| EC50 | 11.0 nM | PMID16257207, PMID15081042 | BindingDB,ChEMBL |
| EC50 | 16.0 nM | PMID23675993 | ChEMBL |
| EC50 | 18.62 nM | None | ChEMBL |
| EC50 | 47.0 nM | PMID27487565 | BindingDB,ChEMBL |
| EC50 | 70.0 nM | PMID18035544 | BindingDB,ChEMBL |
| EC50 | 125.89 nM | PMID18095642 | ChEMBL |
| EC50 | 147.91 nM | PMID23301527 | ChEMBL |
| EC50 | 150.0 nM | PMID23301527 | BindingDB,ChEMBL |
| EC50 | 302.0 nM | PMID25583099 | BindingDB,ChEMBL |
| Efficacy | 98.0 % | PMID16257207 | ChEMBL |
| Emax | 100.0 % | PMID25633969, PMID18035544, PMID19284718 | ChEMBL |
| Kd | 1.25893 nM | PMID8534270 | IUPHAR |
| Ki | 3.98107 - 1000.0 nM | PMID15322733, PMID8534270, PMID12954071, PMID2233697, PMID8114677, PMID8663249 | IUPHAR |
| Ki | 7.77 nM | PMID8632342 | PDSP,BindingDB |
| Ki | 8.2 nM | PMID12954071 | BindingDB |
| Ki | 8.2 nM | PMID16392816, PMID12954071 | PDSP,BindingDB,ChEMBL |
| Ki | 12.02 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 14.0 nM | PMID10611640, PMID16257207, PMID15081042 | PDSP,BindingDB,ChEMBL |
| Ki | 16.2181 nM | PMID15322733 | PDSP |
| Ki | 16.22 nM | PMID15322733 | BindingDB |
| Ki | 21.0 nM | PMID14709324 | PDSP,BindingDB |
| Ki | 30.9 nM | PMID10498829 | BindingDB |
| Ki | 30.903 nM | PMID10498829 | PDSP |
| Ki | 63.09 nM | PMID7582481 | PDSP,BindingDB |
| Ki | 120.0 nM | PMID18468904 | BindingDB,ChEMBL |
| Ki | 130.0 nM | PMID18468904 | BindingDB,ChEMBL |
| Ki | 150.0 nM | PMID18468904 | BindingDB,ChEMBL |
| Ki | 160.0 nM | PMID18468904 | BindingDB,ChEMBL |
| Ki | 174.0 nM | Hoyer et al., PMID1987 | PDSP |
| Ki | 199.52 nM | PMID7582481 | PDSP |
| Ki | 218.77 nM | PMID9225287 | PDSP,BindingDB |
| Ki | 320.0 nM | PMID23301527, PMID11975483 | BindingDB,ChEMBL |
| Ki | 323.59 nM | PMID23301527 | ChEMBL |
| Ki | 341.0 nM | PMID10611640 | PDSP,BindingDB |
| Ki | 398.11 nM | PMID14613313 | ChEMBL |
| Ki | 510.0 nM | PMID8071931 | BindingDB,ChEMBL |
| Ki | 602.55 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 690.0 nM | PMID17067154 | BindingDB,ChEMBL |
| Ki | 3170.75 nM | Andorn et al., PMID1984 | PDSP |
| Ki | 4396.0 nM | Elliot & Kent, PMID1989 | PDSP |
| Relative efficacy | 98.0 % | PMID15081042 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417