

You can:
| Name | 5-hydroxytryptamine receptor 2A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR2A |
| Synonym | 5-HT-2 serotonin receptor 2A serotonin 5HT-2 receptor 5-HT-2A 5-HT2A receptor [ Show all ] |
| Disease | Depression Unspecified Diabetes Erythropoietic porphyria Fibromyalgia [ Show all ] |
| Length | 471 |
| Amino acid sequence | MDILCEENTSLSSTTNSLMQLNDDTRLYSNDFNSGEANTSDAFNWTVDSENRTNLSCEGCLSPSCLSLLHLQEKNWSALLTAVVIILTIAGNILVIMAVSLEKKLQNATNYFLMSLAIADMLLGFLVMPVSMLTILYGYRWPLPSKLCAVWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNSRTKAFLKIIAVWTISVGISMPIPVFGLQDDSKVFKEGSCLLADDNFVLIGSFVSFFIPLTIMVITYFLTIKSLQKEATLCVSDLGTRAKLASFSFLPQSSLSSEKLFQRSIHREPGSYTGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICKESCNEDVIGALLNVFVWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENKKPLQLILVNTIPALAYKSSQLQMGQKKNSKQDAKTTDNDCSMVALGKQHSEEASKDNSDGVNEKVSCV |
| UniProt | P28223 |
| Protein Data Bank | 6a93, 6a94 |
| GPCR-HGmod model | P28223 |
| 3D structure model | This structure is from PDB ID 6a93. |
| BioLiP | BL0441025,BL0441028, BL0441031, BL0441030,BL0441033, BL0441029,BL0441032, BL0441026, BL0441024,BL0441027 |
| Therapeutic Target Database | T32060 |
| ChEMBL | CHEMBL224 |
| IUPHAR | 6 |
| DrugBank | BE0000451 |
| Name | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine |
|---|---|
| Molecular formula | C11H16INO2 |
| IUPAC name | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine |
| Molecular weight | 321.158 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 2.5 |
| Synonyms | AX8051181 DOI-P LS-176383 QC-2023 4-Iodo-2,5-dimethoxy-alpha-methylbenzeneethanamine [ Show all ] |
| Inchi Key | BGMZUEKZENQUJY-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C11H16INO2/c1-7(13)4-8-5-11(15-3)9(12)6-10(8)14-2/h5-7H,4,13H2,1-3H3 |
| PubChem CID | 1229 |
| ChEMBL | CHEMBL6616 |
| IUPHAR | 147 |
| BindingDB | 28582 |
| DrugBank | N/A |
Structure | 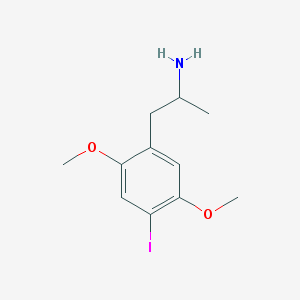 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 6.3 nM | PMID25583099 | BindingDB |
| EC50 | 6.31 nM | PMID25583099 | ChEMBL |
| Emax | 87.0 % | PMID25583099 | ChEMBL |
| Ki | 0.46 nM | PMID9857084 | BindingDB,ChEMBL |
| Ki | 0.631 - 39.8 nM | PMID8534270, PMID2233697, PMID9933142 | IUPHAR |
| Ki | 0.645654 nM | PMID15322733 | PDSP |
| Ki | 0.65 nM | PMID8632342, PMID15322733, PMID12954071 | PDSP,BindingDB |
| Ki | 0.7 nM | PMID9933142 | PDSP,BindingDB |
| Ki | 0.89 nM | PMID10498829 | BindingDB |
| Ki | 0.891251 nM | PMID10498829 | PDSP |
| Ki | 0.92 nM | PMID18847250 | BindingDB,ChEMBL |
| Ki | 0.93 nM | PMID15322733 | BindingDB |
| Ki | 0.933254 nM | PMID15322733 | PDSP |
| Ki | 1.58 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 3.1 nM | PMID10611640 | PDSP,BindingDB |
| Ki | 6.0 nM | Sadzot et al., PMID1989 | PDSP |
| Ki | 13.0 nM | PMID18468904 | BindingDB,ChEMBL |
| Ki | 14.0 nM | PMID18468904 | BindingDB,ChEMBL |
| Ki | 20.47 nM | Pierce & Peroutka, PMID1989 | PDSP |
| Ki | 28.0 nM | PMID18468904 | BindingDB,ChEMBL |
| Ki | 29.0 nM | PMID18468904 | BindingDB,ChEMBL |
| Ki | 38.01 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 52.0 nM | PMID10611640 | PDSP,BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417