| Synonyms | Vospire
SR-01000075161-8
Suxar
Vencronyl
Ventolin (TN)
Salbutamol sulfate
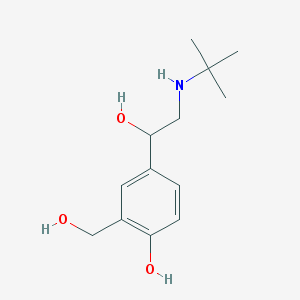
4-[2-(tert-Butylamino)-1-hydroxyethyl]-2-(hydroxymethyl)phenol #
Medolin
CAS-18559-94-9
Salbutamolum
AB00052154-08
NCGC00015955-08
Combivent
Sallbupp
AC1Q1MO0
NCGC00024698-02
Dilatamol
SC-93513
albuterol
NINDS_000943
DTXSID5021255
SPBio_001539
Almotex
Pneumolat
Gerivent
SR-01000075161
AN-18804
Proair HFA
Asmatol
racemic salbutamol
IDI1_000943
BCBcMAP01_000116
Salamol
1,3-Benzenedimethanol, alpha1-(((1,1-dimethylethyl)amino)methyl)-4-hydroxy-
KBioSS_000680
BRD-A88254928-001-04-6
Salbupur
35763-26-9
Lopac0_001098
Bumol
ZX-AFC000812
ST077150
Tox21_110266
Ventamol
Ventoline
Butohaler
Salbutamol, British Pharmacopoeia (BP) Reference Standard
4-{2-[(tert-butyl)amino]-1-hydroxyethyl}-2-(hydroxymethyl)phenol
Mozal
Cetsim
Salbutard
AB00052154_11
NCGC00015955-11
CTK8E5622
Salvent
Aerolin
NCGC00024698-05
DL-N-tert-Butyl-2-(4-hydroxy-3-hydroxymethylphenyl)-2-hydroxyethylamine
SCHEMBL4913
ALBUTEROL SULFATE
NSC757417
EINECS 242-424-0
Spectrum2_001350
alpha-[(t-Butylamino)methyl]-4-hydroxy-m-xylene-alpha,alpha -diol
Prestwick2_000198
HMS1568H17
SR-01000075161-4
API0006973
Proventil HFA
(+-)-Albuterol
HMS3712H17
Asmol
Respolin
.alpha.1-[(tert-Butylamino)methyl]-4-hydroxy-m-xylene-.alpha.,.alpha.'-diol
KBio1_000943
Bio1_000446
Salbron
18559-94-9
KUC106739N
Broncho-Spray
4-Hydroxy-3-hydroxymethyl-alpha-((tert-butylamino)methyl)benzyl alcohol
m-Xylene-.alpha.,.alpha.'-diol, .alpha.'-((tert-butylamino)methyl)-4-hydroxy-
Volma
Sultanol N
to_000081
Ventodisks
4-[2-(tert-butylamino)-1-hydroxy-ethyl]-2-(hydroxymethyl)phenol
m-Xylene-alpha,alpha'-diol, alpha-((tert-butylamino)methyl)-4-hydroxy-
Buventol
Salbutamol,(+/-)
A812971
NCGC00015955-06
CHEMBL714
Salbuven
AC-10286
NCGC00015955-14
SBI-0051060.P005
Airomir
NCGC00258354-01
DSSTox_GSID_21255
SMP1_000268
Albuterol [USAN]
PDSP2_000620
Farcolin
Spectrum_000200
alpha-[(tert-Butylamino)methyl]-4-hydroxy-m-xylene-alpha,alpha'-diol
ProAi (TN)
Asmanil
Q-201696
(alpha1-((tert-butylamino)methyl)-4-hydroxy-m-xylene-alpha,alpha-diol)
HY-B1037
Asthalin (TN)
Sabutal
1,3-Benzenedimethanol, alpha(sup 1)-(((1,1-dimethylethyl)amino)methyl)-4-hydroxy-
KBio2_005816
BIS7017
Salbuhexal
2-t-Butylamino-1-(4-hydroxy-3-hydroxy-3-hydroxymethyl)phenylethanol
Libretin
BSPBio_000155
Salbutamol Free Base
VU0243312-5
ST024764
Theosal
Venetlin (Salt/Mix)
Ventolin Inhaler
Butamol
Salbutamol Sulphate
4-[2-(tert-butylamino)-1-oxidanyl-ethyl]-2-(hydroxymethyl)phenol
MFCD00148978
CC-23825
Salbutamolum [INN-Latin]
AB00052154-09
NCGC00015955-09
CS-4556
Salmaplon
AccuNeb
NCGC00024698-03
DivK1c_000943
SCH-13949W
ALBUTEROL (+/-)
Novosalmol
EC 242-424-0
SPBio_002076
alpha'-((tert-Butylamino)methyl)-4-hydroxy-m-xylene-alpha,alpha'-diol
Prestwick0_000198
Grafalin
SR-01000075161-11
AN-7048
Proventil
HMS2092I08
Asmaven
racemic-Salbutamol
.alpha.'-((tert-Butylamino)methyl)-4-hydroxy-m-xylene-.alpha.,.alpha.'-diol
J10176
BCP28423
Salamol EB
1-(tert-Butylaminomethyl)-4-hydroxy-m-xylene-.alpha.1,.alpha.3-diol
KS-5209
BRD-A88254928-001-05-3
Salbusian
4-(2-(tert-Butylamino)-1-hydroxyethyl)-2-(hydroxymethyl)phenol
LS-29861
ST24042831
Tox21_110266_1
Ventilan
Volare albuterol HFA
m-Xylene-.alpha.,.alpha.-diol, .alpha.-1-((tert-butylamino)methyl)-4-hydroxy-
Butotal
Salbutamol, European Pharmacopoeia (EP) Reference Standard
NCGC00015955-04
CHEBI:2549
Salbutine
AB0011383
NCGC00015955-12
D02147
Saventol
Aerolin (TN)
NCGC00024698-06
dl-Salbutamol
Servitamol
Albuterol Sulphate
Parasma
Eolene
Spectrum4_000887
alpha-[(t-Butylamino)methyl]-4-hydroxy-m-xylene-alpha,alpha'-diol
Prestwick3_000198
HMS1921C04
Arubendol-Salbutamol
Proventil Inhaler
(+-)-alpha(sup 1)-(((1,1-Dimethylethyl)amino)methyl)-4-hydroxy-1,3-benzenedimethanol
HMS502P05
Asmol Uni-Dose
Rotahaler
1,3-Benzenedimethanol, .alpha.(1)-[[(1,1-dimethylethyl)amino]methyl]-4-hydroxy-
KBio2_000680
Bio1_000935
Salbu-BASF
2-(tert-Butylamino)-1-(4-hydroxy-3-hydroxymethylphenyl)ethanol
L000531
Broncovaleas
Salbutamol (Albuterol)
4-[(1S)-1-hydroxy-2-(propylamino)ethyl]-2-(hydroxymethyl)phenol, oxamethane
Volmax
SR-01000075161-5
Suprasma
TR-008568
Ventolin
4-[2-(tert-Butylamino)-1-hydroxyethyl]-2-(hydroxymethyl)phenol
MCULE-4493408325
C-14301
Salbutamol-d9
AB00052154
NCGC00015955-07
Cobutolin
Salbuvent
AC1L1CVO
NCGC00015955-16
DB01001
SC-22638
AKOS007930174
NDAUXUAQIAJITI-UHFFFAOYSA-N
DSSTox_RID_76042
Solbutamol
Albuterol, United States Pharmacopeia (USP) Reference Standard
Pharmakon1600-01500677
FT-0630453
Spreor
Alti-Salbutamol
ProAir
Asmasal
R,S-Albuterol
(l)-albuterol
I06-0331
AX8136202
Sabutamol
1,3-Benzenedimethanol, alpha(sup 1)-(((1,1-dimethylethyl)amino)methyl)-4-hydroxy-, (+-)-
KBioGR_001294
BPBio1_000171
Salbulin
Lopac0_001090
Bugonol
Salbutamol impurity I (1RS)-2-[(1,1-dimethylethyl)
Zaperin
ST075204
Tobybron
Ventalin inhaler
Ventolin Rotacaps
Buto-Asma
Salbutamol [INN:BAN]
4-{2-[(1,1-dimethylethyl)amino]-1-hydroxyethyl}-2-(hydroxymethyl)phenol
MolPort-001-838-925
CCG-39288
Salbutan
AB00052154_10
NCGC00015955-10
CS0015
Salomol
ACM35763269
NCGC00024698-04
dl-Albuterol
SCHEMBL10025126
Albuterol (USP)
NSC-757417
Ecovent
SPECTRUM1500677
alpha(sup 1)-((tert-Butylamino)methyl)-4-hydroxy-m-xylene-alpha,alpha'-diol
Prestwick1_000198
GTPL558
SR-01000075161-3
Anebron
Proventil (TN)
HMS2095H17
Asmidon
Respax
.alpha.1'-[[(1,1-Dimethylethyl)amino]methyl]-4-hydroxy-1,3-benzenedimethanol, hemisulfate
K-5595
BDBM25769
Salbetol
1-(tert-Butylaminomethyl)-4-hydroxy-m-xylene-alpha1,alpha3-diol
KSC-11-222-1
BRN 2213614
Salbutalan
4-Hydroxy-3-hydroxymethyl-.alpha.-((tert-butylamino)methyl)benzyl alcohol
LS-7693
Sultanol
Tox21_200800
Ventiloboi
Volare easi-breathe
4-[(1S)-2-(butylamino)-1-hydroxy-isopropyl]-2-(hydroxymethyl)phenol
m-Xylene-alpha,alpha'-diol, alpha'-((tert-butylamino)methyl)-4-hydroxy-
Butovent
Salbutamol, VETRANAL(TM), analytical standard
559S949
NCGC00015955-05
Salbutol
AB1006817
NCGC00015955-13
D0K5CB
SBB066105
AH 3365
NCGC00024698-07
DSSTox_CID_1255
SGCUT00011
Albuterol [USAN:USP]
PDSP1_000623
EU-0101090
Spectrum5_001059
Alpha-[(t-Butylamino)methyl]-4-hydroxy-m-xylene-alpha,alpha-diol
Prestwick_665
HMS2089I19
Asmadil
Q-201695
(+-)-Salbutamol
HSDB 7206
Asthalin
S 8260
1,3-Benzenedimethanol, .alpha.1-[[(1,1-dimethylethyl)amino]methyl]-4-hydroxy-
KBio2_003248
Bio1_001424
Salbu-Fatol
2-(tert-Butylamino)-1-[4-hydroxy-3-(hydroxymethyl)phenyl]ethanol
Levosalbutamol Hydrochloride
Bronter
Salbutamol 100 microg/mL in Acetonitrile [ Show all ] |
|---|


![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417