

You can:
| Name | Cgs 15943 |
|---|---|
| Molecular formula | C13H8ClN5O |
| IUPAC name | 9-chloro-2-(furan-2-yl)-[1,2,4]triazolo[1,5-c]quinazolin-5-amine |
| Molecular weight | 285.691 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 1 |
| XlogP | 2.5 |
| Synonyms | Tox21_500324 9-chloro-2-(furan-2-yl)[1,2,4]triazolo[1,5-c]quinazolin-5-amine AKOS015906465 CCG-100792 HMS2051C13 [ Show all ] |
| Inchi Key | MSJODEOZODDVGW-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C13H8ClN5O/c14-7-3-4-9-8(6-7)12-17-11(10-2-1-5-20-10)18-19(12)13(15)16-9/h1-6H,(H2,15,16) |
| PubChem CID | 2690 |
| ChEMBL | CHEMBL16687 |
| IUPHAR | 384 |
| BindingDB | 50004566 |
| DrugBank | N/A |
Structure | 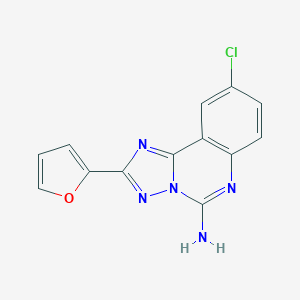 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| ED50 | 0.3 uM | PMID8676354 | ChEMBL |
| IC50 | 12.0 nM | PMID9622554 | BindingDB,ChEMBL |
| Kd | 3.242 nM | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| Ki | 0.14 nM | PMID9258366 | BindingDB |
| Ki | 0.142 nM | PMID9258366 | ChEMBL |
| Ki | 0.35 nM | PMID14584936, PMID15734651 | BindingDB,ChEMBL |
| Ki | 0.398107 - 19.9526 nM | PMID9459566, PMID9933143, PMID7775460, PMID9179373 | IUPHAR |
| Ki | 0.4 nM | PMID18637670, PMID9933143 | PDSP,BindingDB,ChEMBL |
| Ki | 0.41 nM | PMID14584936 | BindingDB,ChEMBL |
| Ki | 0.43 nM | PMID11123985 | BindingDB,ChEMBL |
| Ki | 0.45 nM | PMID15734651 | BindingDB |
| Ki | 0.457 nM | PMID9258366 | ChEMBL |
| Ki | 0.46 nM | PMID9258366 | BindingDB |
| Ki | 0.52 nM | PMID14584936 | BindingDB,ChEMBL |
| Ki | 0.67 nM | PMID14584936 | ChEMBL |
| Ki | 0.67 nM | PMID14584936 | BindingDB |
| Ki | 0.84 nM | PMID9258366, PMID15734651, PMID14584936 | BindingDB,ChEMBL |
| Ki | 0.84 nM | PMID14584936 | BindingDB |
| Ki | 0.842 nM | PMID9258366 | ChEMBL |
| Ki | 0.91 nM | PMID14584936, PMID15734651 | BindingDB,ChEMBL |
| Ki | 0.91 nM | PMID14584936 | BindingDB |
| Ki | 1.0 nM | PMID14584936 | BindingDB,ChEMBL |
| Ki | 1.7 nM | PMID8863790 | BindingDB,ChEMBL |
| Ki | 2.27 nM | PMID20202853 | BindingDB,ChEMBL |
| Ki | 4.18 nM | PMID9459566 | PDSP,BindingDB |
| kon | 370200.0 Ms-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| k_off | 0.0012 s-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417