

You can:
| Name | 2-oxoglutarate receptor 1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Oxgr1 |
| Synonym | P2Y purinoceptor 15 P2RY15 oxoglutarate receptor GPR99 GPR80 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 337 |
| Amino acid sequence | MIETLDSPANDSDFLDYITALENCTDEQISFKMQYLPVIYSIIFLVGFPGNTVAISIYVFKMRPWKSSTIIMLNLALTDLLYLTSLPFLIHYYASGENWIFGDFMCKFIRFGFHFNLYSSILFLTCFSLFRYIVIIHPMSCFSIQKTRWAVVACAGVWVISLVAVMPMTFLITSTTRTNRSACLDLTSSDDLTTIKWYNLILTATTFCLPLLIVTLCYTTIISTLTHGPRTHSCFKQKARRLTILLLLVFYVCFLPFHILRVIRIESRLLSISCSIESHIHEAYIVSRPLAALNTFGNLLLYVVVSNNFQQAFCSAVRCKAIGDLEQAKKDSCSNNP |
| UniProt | Q6Y1R5 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2325 |
| IUPHAR | N/A |
| DrugBank | N/A |
| Name | CHEMBL608003 |
|---|---|
| Molecular formula | C19H22N6O6 |
| IUPAC name | (2S,3S,4R)-5-(6-aminopurin-9-yl)-N-[(3,4-dimethoxyphenyl)methyl]-3,4-dihydroxyoxolane-2-carboxamide |
| Molecular weight | 430.421 |
| Hydrogen bond acceptor | 10 |
| Hydrogen bond donor | 4 |
| XlogP | -0.5 |
| Synonyms | BDBM50369928 |
| Inchi Key | LQAZHNHWXDUXTM-FRLFKWGPSA-N |
| Inchi ID | InChI=1S/C19H22N6O6/c1-29-10-4-3-9(5-11(10)30-2)6-21-18(28)15-13(26)14(27)19(31-15)25-8-24-12-16(20)22-7-23-17(12)25/h3-5,7-8,13-15,19,26-27H,6H2,1-2H3,(H,21,28)(H2,20,22,23)/t13-,14+,15-,19?/m0/s1 |
| PubChem CID | 46875320 |
| ChEMBL | CHEMBL608003 |
| IUPHAR | N/A |
| BindingDB | 50369928 |
| DrugBank | N/A |
Structure | 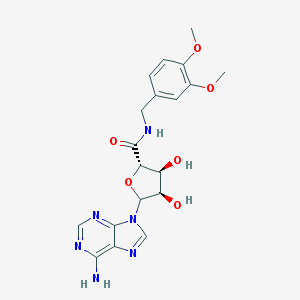 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | 140.0 nM | PMID11170630 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417