

You can:
| Name | 5-hydroxytryptamine receptor 2A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR2A |
| Synonym | 5-HT-2 serotonin receptor 2A serotonin 5HT-2 receptor 5-HT-2A 5-HT2A receptor [ Show all ] |
| Disease | Depression Unspecified Diabetes Erythropoietic porphyria Fibromyalgia [ Show all ] |
| Length | 471 |
| Amino acid sequence | MDILCEENTSLSSTTNSLMQLNDDTRLYSNDFNSGEANTSDAFNWTVDSENRTNLSCEGCLSPSCLSLLHLQEKNWSALLTAVVIILTIAGNILVIMAVSLEKKLQNATNYFLMSLAIADMLLGFLVMPVSMLTILYGYRWPLPSKLCAVWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNSRTKAFLKIIAVWTISVGISMPIPVFGLQDDSKVFKEGSCLLADDNFVLIGSFVSFFIPLTIMVITYFLTIKSLQKEATLCVSDLGTRAKLASFSFLPQSSLSSEKLFQRSIHREPGSYTGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICKESCNEDVIGALLNVFVWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENKKPLQLILVNTIPALAYKSSQLQMGQKKNSKQDAKTTDNDCSMVALGKQHSEEASKDNSDGVNEKVSCV |
| UniProt | P28223 |
| Protein Data Bank | 6a93, 6a94 |
| GPCR-HGmod model | P28223 |
| 3D structure model | This structure is from PDB ID 6a93. |
| BioLiP | BL0441025,BL0441028, BL0441031, BL0441030,BL0441033, BL0441029,BL0441032, BL0441026, BL0441024,BL0441027 |
| Therapeutic Target Database | T32060 |
| ChEMBL | CHEMBL224 |
| IUPHAR | 6 |
| DrugBank | BE0000451 |
| Name | haloperidol |
|---|---|
| Molecular formula | C21H23ClFNO2 |
| IUPAC name | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one |
| Molecular weight | 375.868 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 1 |
| XlogP | 3.2 |
| Synonyms | ZINC537822 4763-EP2277876A1 DB00502 4763-EP2305260A1 DTXSID4034150 [ Show all ] |
| Inchi Key | LNEPOXFFQSENCJ-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C21H23ClFNO2/c22-18-7-5-17(6-8-18)21(26)11-14-24(15-12-21)13-1-2-20(25)16-3-9-19(23)10-4-16/h3-10,26H,1-2,11-15H2 |
| PubChem CID | 3559 |
| ChEMBL | CHEMBL54 |
| IUPHAR | 86 |
| BindingDB | 21398 |
| DrugBank | DB00502 |
Structure | 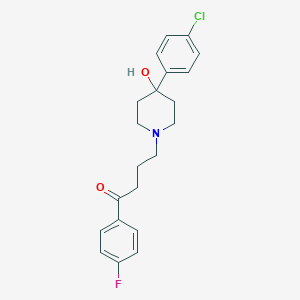 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Affinity ratio | 0.016 - | PMID8568801 | ChEMBL |
| IC50 | 75.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| IC50 | 288.0 nM | PMID8831770 | BindingDB,ChEMBL |
| Ki | 21.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Ki | 25.0 nM | PMID9015795, PMID14998318 | PDSP,BindingDB,ChEMBL |
| Ki | 30.0 nM | PMID16051647 | PDSP |
| Ki | 36.0 nM | Wander et al., PMID1987 | PDSP |
| Ki | 46.0 nM | PMID9577836, http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6W84-42KD8D9-7&_coverDate=01%2F31%2F2001&_alid=93617739&_rdoc=1&_fmt=&_orig=search&_qd=1&_cdi=6644&_sort=d&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=5e5748d1ff8f5317fb6669f7ddc989f6 | PDSP,BindingDB |
| Ki | 50.1187 - 199.526 nM | PMID8935801, PMID9732398, PMID12629531, PMID15102927, PMID18308814 | IUPHAR |
| Ki | 53.0 nM | PMID12629531 | PDSP,BindingDB |
| Ki | 53.31 nM | Andorn et al., PMID1984 | PDSP |
| Ki | 60.0 nM | http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6W84-42KD8D9-7&_coverDate=01%2F31%2F2001&_alid=93617739&_rdoc=1&_fmt=&_orig=search&_qd=1&_cdi=6644&_sort=d&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=5e5748d1ff8f5317fb6669f7ddc989f6 | PDSP |
| Ki | 61.0 nM | PMID11132243 | PDSP,BindingDB |
| Ki | 70.2 nM | PMID15102927 | PDSP,BindingDB |
| Ki | 75.85 nM | PMID9225287 | PDSP,BindingDB |
| Ki | 100.0 nM | , None, PMID17880057 | BindingDB,ChEMBL |
| Ki | 103.0 nM | http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6W84-42KD8D9-7&_coverDate=01%2F31%2F2001&_alid=93617739&_rdoc=1&_fmt=&_orig=search&_qd=1&_cdi=6644&_sort=d&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=5e5748d1ff8f5317fb6669f7ddc989f6 | PDSP |
| Ki | 108.0 nM | PMID8831770 | BindingDB,ChEMBL |
| Ki | 112.0 nM | PMID7520908 | PDSP,BindingDB |
| Ki | 120.0 nM | PMID18595716, PMID25070422 | PDSP,BindingDB,ChEMBL |
| Ki | 130.0 nM | PMID14695828 | PDSP,BindingDB,ChEMBL |
| Ki | 165.96 nM | PMID18783204, PMID17588750, PMID24316025, PMID19796944, PMID14741248, MedChemComm, (2011) 2:12:1194 | BindingDB,ChEMBL |
| Ki | 166.0 nM | PMID14741248 | BindingDB |
| Ki | 186.0 nM | PMID8997630 | PDSP,BindingDB |
| Ki | 200.0 nM | PMID8935801 | PDSP,BindingDB |
| Ki | 435.0 nM | PMID15771415 | PDSP,BindingDB,ChEMBL |
| Ratio | 0.82 - | PMID11784139 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417