| Name | Adenosine receptor A2a |
|---|
| Species | Homo sapiens (Human) |
|---|
| Gene | ADORA2A |
|---|
| Synonym | RDC8
adenosine receptor A2a
A2A receptor
A2-AR |
|---|
| Disease | Radionuclide imaging
Diabetic foot ulcer
Glaucoma
Hypertension
Neuropathic pain
Pain
Coronary disorder diagnosis
Parkinson's disease
Allergic rhinitis
Arteriosclerosis
Asthma; Chronic obstructive pulmonary disease [ Show all ] |
|---|
| Length | 412 |
|---|
| Amino acid sequence | MPIMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKNHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESQPLPGERARSTLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPFKAAGTSARVLAAHGSDGEQVSLRLNGHPPGVWANGSAPHPERRPNGYALGLVSGGSAQESQGNTGLPDVELLSHELKGVCPEPPGLDDPLAQDGAGVS |
|---|
| UniProt | P29274 |
|---|
| Protein Data Bank | 4ug2, 4eiy, 3vga, 3vg9, 3uzc, 3uza, 3rfm, 3rey, 3pwh, 5iu4, 3eml, 2ydv, 2ydo, 3qak, 4uhr, 5g53, 5olh, 5olo, 5olv, 5olz, 5om1, 5om4, 5uvi, 5vra, 5wf5, 5wf6, 6aqf, 6gdg, 5olg, 5nm4, 5iu7, 5iu8, 5iua, 5iub, 5jtb, 5k2a, 5k2b, 5mzj, 5mzp, 5n2r, 5nlx, 5nm2 |
|---|
| GPCR-HGmod model | P29274 |
|---|
| 3D structure model | This structure is from PDB ID 4ug2.
|
|---|
| BioLiP | BL0213760, BL0350712, BL0350713,BL0350714,BL0350715, BL0353317,BL0353318, BL0357562, BL0357563,BL0357564,BL0357565, BL0357566, BL0357567,BL0357568,BL0357569, BL0379362,BL0379363,BL0379364, BL0379725, BL0379726,BL0379727,BL0379728, BL0379729, BL0385550, BL0350708,BL0350709,BL0350710,, BL0350707, BL0350703,BL0350704,BL0350705,, BL0215974, BL0215975, BL0227995, BL0227996, BL0227997,BL0227998,BL0227999, BL0312021,BL0312022, BL0312023, BL0350692, BL0350693,BL0350694,BL0350695,, BL0350697, BL0350698,BL0350699,BL0350700,, BL0350702, BL0385551,BL0385552,BL0385553, BL0385557, BL0385558,BL0385559,BL0385560,, BL0401931,BL0401932,BL0401933, BL0401934, BL0401935,BL0401936,BL0401937, BL0401938, BL0401939,BL0401940,BL0401941,, BL0401943, BL0401944,BL0401945,BL0401946,, BL0401948, BL0401949,BL0401950,BL0401951,, BL0401954,BL0401955,BL0401956,, BL0405662, BL0405663, BL0401930, BL0401927,BL0401928,BL0401929, BL0401926, BL0385572, BL0385573,BL0385574,BL0385575, BL0393144, BL0393145, BL0393146, BL0393147,BL0393148,BL0393149, BL0393150, BL0393151,BL0393152, BL0398902, BL0398903,BL0398904,BL0398905, BL0401593, BL0401594,BL0401595,BL0401596, BL0414567, BL0213751, BL0401953, BL0379361, BL0130764, BL0130785, BL0152618, BL0194187, BL0195884, BL0199981, BL0200022 |
|---|
| Therapeutic Target Database | T77365 |
|---|
| ChEMBL | CHEMBL251 |
|---|
| IUPHAR | 19 |
|---|
| DrugBank | BE0000924 |
|---|


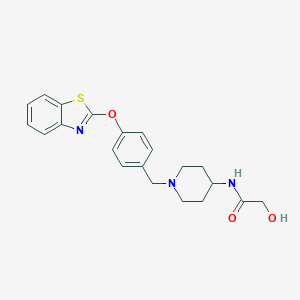
![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417