

You can:
| Name | Histamine H4 receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Hrh4 |
| Synonym | GPCR105 H4 receptor H4R HH4R |
| Disease | N/A for non-human GPCRs |
| Length | 391 |
| Amino acid sequence | MSESNGTDVLPLTAQVPLAFLMSLLAFAITIGNAVVILAFVADRNLRHRSNYFFLNLAISDFFVGVISIPLYIPHTLFNWNFGSGICMFWLITDYLLCTASVYSIVLISYDRYQSVSNAVRYRAQHTGILKIVAQMVAVWILAFLVNGPMILASDSWKNSTNTEECEPGFVTEWYILAITAFLEFLLPVSLVVYFSVQIYWSLWKRGSLSRCPSHAGFIATSSRGTGHSRRTGLACRTSLPGLKEPAASLHSESPRGKSSLLVSLRTHMSGSIIAFKVGSFCRSESPVLHQREHVELLRGRKLARSLAVLLSAFAICWAPYCLFTIVLSTYRRGERPKSIWYSIAFWLQWFNSLINPFLYPLCHRRFQKAFWKILCVTKQPAPSQTQSVSS |
| UniProt | Q91ZY1 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4468 |
| IUPHAR | 265 |
| DrugBank | N/A |
| Name | JNJ-7777120 |
|---|---|
| Molecular formula | C14H16ClN3O |
| IUPAC name | (5-chloro-1H-indol-2-yl)-(4-methylpiperazin-1-yl)methanone |
| Molecular weight | 277.752 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 1 |
| XlogP | 2.3 |
| Synonyms | MCULE-4664243664 RT-013403 [3H]-JNJ7777120 (5-Chloro-1H-indol-2-yl)-(4-methyl-piperazin-1-yl)-methanone 2490AH [ Show all ] |
| Inchi Key | HUQJRYMLJBBEDO-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C14H16ClN3O/c1-17-4-6-18(7-5-17)14(19)13-9-10-8-11(15)2-3-12(10)16-13/h2-3,8-9,16H,4-7H2,1H3 |
| PubChem CID | 4908365 |
| ChEMBL | CHEMBL129198 |
| IUPHAR | 1279, 1278 |
| BindingDB | 22566 |
| DrugBank | N/A |
Structure | 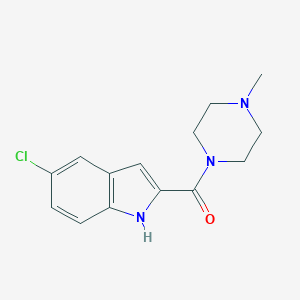 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 2.57 nM | PMID18817367 | ChEMBL |
| EC50 | 42.66 nM | PMID18811133 | ChEMBL |
| Efficacy | 23.0 % | PMID18811133 | ChEMBL |
| Ki | 2.4 nM | PMID12954048 | BindingDB,ChEMBL |
| Ki | 2.6 nM | PMID14722321 | BindingDB |
| Ki | 2.692 nM | PMID18817367, PMID18811133 | ChEMBL |
| Ki | 2.7 nM | PMID18817367, PMID18811133 | BindingDB |
| Ki | 4.266 nM | PMID21920751 | ChEMBL |
| Ki | 4.27 nM | PMID21920751 | BindingDB |
| Ki | 4.677 nM | PMID18983139 | ChEMBL |
| Ki | 4.7 nM | PMID18983139 | BindingDB |
| Ki | 6.0 nM | PMID22153663 | BindingDB,ChEMBL |
| pKb | 7.19 - | PMID18811133 | ChEMBL |
| pKb | 8.51 - | PMID18983139 | ChEMBL |
| pKb | 8.59 - | PMID18811133 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417