

You can:
| Name | Cannabinoid receptor 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CNR2 |
| Synonym | Peripheral cannabinoid receptor rCB2 hCB2 cannabinoid receptor 2 (macrophage) cannabinoid receptor 2 (spleen) [ Show all ] |
| Disease | Immune disorder Inflammatory bowel disease Inflammatory disease Neuropathic pain Osteoporosis [ Show all ] |
| Length | 360 |
| Amino acid sequence | MEECWVTEIANGSKDGLDSNPMKDYMILSGPQKTAVAVLCTLLGLLSALENVAVLYLILSSHQLRRKPSYLFIGSLAGADFLASVVFACSFVNFHVFHGVDSKAVFLLKIGSVTMTFTASVGSLLLTAIDRYLCLRYPPSYKALLTRGRALVTLGIMWVLSALVSYLPLMGWTCCPRPCSELFPLIPNDYLLSWLLFIAFLFSGIIYTYGHVLWKAHQHVASLSGHQDRQVPGMARMRLDVRLAKTLGLVLAVLLICWFPVLALMAHSLATTLSDQVKKAFAFCSMLCLINSMVNPVIYALRSGEIRSSAHHCLAHWKKCVRGLGSEAKEEAPRSSVTETEADGKITPWPDSRDLDLSDC |
| UniProt | P34972 |
| Protein Data Bank | 5zty |
| GPCR-HGmod model | P34972 |
| 3D structure model | This structure is from PDB ID 5zty. |
| BioLiP | BL0438927 |
| Therapeutic Target Database | T37693 |
| ChEMBL | CHEMBL253 |
| IUPHAR | 57 |
| DrugBank | BE0000095 |
| Name | Win-55212-2 |
|---|---|
| Molecular formula | C27H26N2O3 |
| IUPAC name | [(11R)-2-methyl-11-(morpholin-4-ylmethyl)-9-oxa-1-azatricyclo[6.3.1.04,12]dodeca-2,4(12),5,7-tetraen-3-yl]-naphthalen-1-ylmethanone |
| Molecular weight | 426.516 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 0 |
| XlogP | 4.4 |
| Synonyms | NCGC00016210-06 SCHEMBL118531 (r)-(+)-[2,3-dihydro-5-methyl-3-(4-morpholinylmethyl)pyrrolo[1,2,3-de]-1,4-benzoxazin-6-yl]-1-naphthalenylmethanone CHEBI:73295 HQVHOQAKMCMIIM-HXUWFJFHSA-N [ Show all ] |
| Inchi Key | HQVHOQAKMCMIIM-HXUWFJFHSA-N |
| Inchi ID | InChI=1S/C27H26N2O3/c1-18-25(27(30)22-9-4-7-19-6-2-3-8-21(19)22)23-10-5-11-24-26(23)29(18)20(17-32-24)16-28-12-14-31-15-13-28/h2-11,20H,12-17H2,1H3/t20-/m1/s1 |
| PubChem CID | 5311501 |
| ChEMBL | CHEMBL188 |
| IUPHAR | 733 |
| BindingDB | 21281 |
| DrugBank | N/A |
Structure | 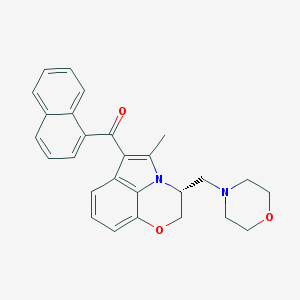 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 50.0 % | PMID19091565 | ChEMBL |
| Activity | 74.0 % | PMID19921781, PMID18311894 | ChEMBL |
| Activity | 98.0 % | PMID19921781 | ChEMBL |
| Activity | 127.8 % | PMID16392793 | ChEMBL |
| Activity | 207.1 % | PMID17915849 | ChEMBL |
| EC50 | 0.39 nM | PMID19921781 | BindingDB,ChEMBL |
| EC50 | 0.47 nM | PMID17630726 | BindingDB,ChEMBL |
| EC50 | 1.5 nM | PMID17884494 | BindingDB,ChEMBL |
| EC50 | 1.585 nM | PMID21885167 | ChEMBL |
| EC50 | 2.0 nM | PMID24508127 | ChEMBL |
| EC50 | 2.04 nM | PMID23849204 | ChEMBL |
| EC50 | 14.0 nM | PMID18522867 | BindingDB,ChEMBL |
| EC50 | 14.8 nM | PMID23350768 | ChEMBL |
| EC50 | 15.0 nM | PMID23350768 | BindingDB |
| EC50 | 15.6 nM | PMID26922225 | ChEMBL |
| EC50 | 16.0 nM | PMID26922225 | BindingDB |
| EC50 | 24.5 nM | PMID25699149 | ChEMBL |
| EC50 | 24.6 nM | PMID16392793, PMID20979417 | BindingDB,ChEMBL |
| EC50 | 25.0 nM | PMID25699149 | BindingDB |
| EC50 | 86.0 nM | PMID19921781, PMID18311894 | BindingDB,ChEMBL |
| EC50 | 89.6 nM | PMID23017078, PMID23151320 | BindingDB,ChEMBL |
| EC50 | 124.5 nM | PMID18311894 | BindingDB |
| Emax | 53.0 % | PMID26922225 | ChEMBL |
| Emax | 64.0 % | PMID18522867 | ChEMBL |
| Emax | 96.9 % | PMID17630726 | ChEMBL |
| Emax | 100.0 % | PMID17884494 | ChEMBL |
| Emax | 126.2 % | PMID16392793 | ChEMBL |
| Emax | 132.0 % | PMID26922225 | ChEMBL |
| Emax | 207.0 % | PMID25699149 | ChEMBL |
| Emax | 207.1 % | PMID20979417 | ChEMBL |
| Emax | 232.0 % | PMID23017078, PMID23151320 | ChEMBL |
| Emax | 243.0 % | PMID23849204, PMID24508127 | ChEMBL |
| IC50 | 1.1 nM | PMID17521907 | BindingDB,ChEMBL |
| IC50 | 3.4 nM | PMID23227781 | BindingDB,ChEMBL |
| IC50 | 8.9 nM | PMID17630726 | BindingDB,ChEMBL |
| IC50 | 15.7 nM | PMID22738271 | BindingDB,ChEMBL |
| Inhibition | 70.0 % | PMID26789378 | ChEMBL |
| Intrinsic activity | 190.0 % | PMID24922543 | ChEMBL |
| Ki | 0.251188 - 3.98108 nM | PMID7565624, PMID8819477, PMID8679694 | IUPHAR |
| Ki | 0.28 nM | PMID22341572, PMID16005223, PMID27560280, PMID9857088 | BindingDB,ChEMBL |
| Ki | 0.28 nM | PMID27560280 | BindingDB |
| Ki | 0.29 nM | PMID11160626 | BindingDB |
| Ki | 0.3 nM | PMID12747783, PMID23865723, PMID23466226, PMID18680277, PMID23672690 | BindingDB,ChEMBL |
| Ki | 0.3 nM | PMID23865723, PMID23466226 | BindingDB |
| Ki | 0.41 nM | PMID12065738 | BindingDB |
| Ki | 0.45 nM | PMID24831513, PMID25072877 | BindingDB,ChEMBL |
| Ki | 0.45 nM | PMID25072877 | BindingDB |
| Ki | 0.9 nM | PMID26149623 | BindingDB |
| Ki | 0.9 nM | PMID26149623 | ChEMBL |
| Ki | 1.19 nM | PMID10688601 | BindingDB |
| Ki | 1.2 nM | PMID19278853 | BindingDB,ChEMBL |
| Ki | 1.288 nM | PMID19921781, PMID18311894 | ChEMBL |
| Ki | 1.3 nM | PMID19921781, PMID18311894, PMID21183257 | BindingDB,ChEMBL |
| Ki | 2.0 nM | PMID22044209, PMID16213718, PMID22607668 | BindingDB,ChEMBL |
| Ki | 2.1 nM | PMID25065940, PMID21999614, PMID21902175, PMID16279794, PMID19331413, PMID17942307, PMID17561406, PMID27448919, PMID20022504 | BindingDB,ChEMBL |
| Ki | 2.2 nM | PMID17521907 | BindingDB,ChEMBL |
| Ki | 2.51 nM | PMID21316962 | BindingDB |
| Ki | 2.512 nM | MedChemComm, (2010) 1:1:54, PMID21316962, PMID21074434 | ChEMBL |
| Ki | 2.9 nM | PMID17884494 | BindingDB,ChEMBL |
| Ki | 3.1 nM | PMID25699149, PMID23017078, PMID23151320 | BindingDB,ChEMBL |
| Ki | 3.7 nM | PMID26789378, PMID26209834, PMID24141201, PMID24378710, PMID23434135 | BindingDB,ChEMBL |
| Ki | 3.73 nM | PMID23434135 | ChEMBL |
| Ki | 3.9 nM | PMID11741470 | BindingDB,ChEMBL |
| Ki | 4.0 nM | PMID12161142 | BindingDB,ChEMBL |
| Ki | 4.53 nM | PMID22738271 | BindingDB,ChEMBL |
| Ki | 4.6 nM | PMID26922225 | BindingDB,ChEMBL |
| Ki | 4.95 nM | PMID23350768 | ChEMBL |
| Ki | 5.0 nM | PMID23350768 | BindingDB |
| Ki | 6.31 nM | PMID21885167 | ChEMBL |
| Ki | 9.1 nM | PMID16392793, PMID24508127, PMID23849204, PMID20979417, PMID20688519 | BindingDB,ChEMBL |
| Ki | 13.0 nM | PMID24125850 | ChEMBL |
| Ki | 16.1 nM | PMID23849204 | ChEMBL |
| Ki | 20.0 nM | PMID18522867 | BindingDB,ChEMBL |
| Ki | 70.0 nM | PMID24139843 | ChEMBL |
| Log Ki | 0.55 nM | PMID10882356 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417