

You can:
| Name | Kappa-type opioid receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | OPRK1 |
| Synonym | K-OR-1 KOPr OP2 KOP KOR-1 [ Show all ] |
| Disease | Obesity Opiate dependence Inflammatory bowel disease Erythema Diarrhea-predominant IBS [ Show all ] |
| Length | 380 |
| Amino acid sequence | MDSPIQIFRGEPGPTCAPSACLPPNSSAWFPGWAEPDSNGSAGSEDAQLEPAHISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVISIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGSREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSTSHSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFPLKMRMERQSTSRVRNTVQDPAYLRDIDGMNKPV |
| UniProt | P41145 |
| Protein Data Bank | 6b73, 4djh |
| GPCR-HGmod model | P41145 |
| 3D structure model | This structure is from PDB ID 6b73. |
| BioLiP | BL0402244,BL0402246, BL0224693,BL0224694, BL0402243,BL0402245 |
| Therapeutic Target Database | T60693 |
| ChEMBL | CHEMBL237 |
| IUPHAR | 318 |
| DrugBank | BE0000632 |
| Name | Norbinaltorphimine |
|---|---|
| Molecular formula | C40H43N3O6 |
| IUPAC name | (1S,2S,7S,8S,12R,20R,24R,32R)-11,33-bis(cyclopropylmethyl)-19,25-dioxa-11,22,33-triazaundecacyclo[24.9.1.18,14.01,24.02,32.04,23.05,21.07,12.08,20.030,36.018,37]heptatriaconta-4(23),5(21),14(37),15,17,26,28,30(36)-octaene-2,7,17,27-tetrol |
| Molecular weight | 661.799 |
| Hydrogen bond acceptor | 8 |
| Hydrogen bond donor | 5 |
| XlogP | 3.2 |
| Synonyms | NCGC00024547-02 11,12,13,14,19a,20b-dodecahydro-, (4bS,8R,8aS,10aS,11R,14aS,19aR,20bR)- 4,8:11,15-Dimethano-20H-bisbenzofuro[2,3-a:3',2'-i]dipyrido[4,3-b:3',4'-h]carbazole-1,8a,10a,18-tetrol, 7,12-bis(cyclopropylmethyl)-5,6,7,8,9,10,11,12,13,14,19a,20b-dodecahydro-, (4bS,8R,8aS,10aS,11R, CHEBI:81529 D05QJS [ Show all ] |
| Inchi Key | APSUXPSYBJVPPS-YAUKWVCOSA-N |
| Inchi ID | InChI=1S/C40H43N3O6/c44-25-7-5-21-13-27-39(46)15-23-24-16-40(47)28-14-22-6-8-26(45)34-30(22)38(40,10-12-43(28)18-20-3-4-20)36(49-34)32(24)41-31(23)35-37(39,29(21)33(25)48-35)9-11-42(27)17-19-1-2-19/h5-8,19-20,27-28,35-36,41,44-47H,1-4,9-18H2/t27-,28-,35+,36+,37+,38+,39-,40-/m1/s1 |
| PubChem CID | 5480230 |
| ChEMBL | CHEMBL573214 |
| IUPHAR | 1642 |
| BindingDB | 82551 |
| DrugBank | N/A |
Structure | 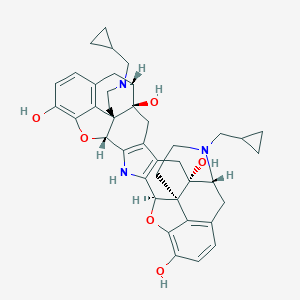 |
| Lipinski's druglikeness | This ligand is heavier than 500 daltons. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 50.0 % | PMID20568781 | ChEMBL |
| EC50 | 0.039 Ke nM-1 | PMID12672258 | ChEMBL |
| EC50 | 0.039 nM | PMID12672258 | BindingDB |
| EC50 | 6.0 nM | PMID20568781 | BindingDB,ChEMBL |
| IC50 | 0.28 nM | PMID25593096 | ChEMBL |
| IC50 | 0.28 nM | PMID25593096 | BindingDB |
| IC50 | 2.1 nM | PMID20727752, PMID20719509 | BindingDB,ChEMBL |
| IC50 | 2.5 nM | PMID25593096 | BindingDB,ChEMBL |
| IC50 | 4.37 nM | PMID12747782 | ChEMBL |
| IC50 | 4.4 nM | PMID12747782 | BindingDB |
| IC50 | 4.6 nM | PMID25593096 | BindingDB,ChEMBL |
| Inhibition | <86.0 % | PMID25593096 | ChEMBL |
| Kb | 0.798 nM | PMID21958337 | ChEMBL |
| Ke | 0.03 nM | PMID15456250 | ChEMBL |
| Ke | 0.039 nM | PMID24973818 | ChEMBL |
| Ke | 0.04 nM | PMID12519069, PMID12139463, PMID14640558, PMID23360448, PMID16366600, PMID12825951, PMID12723940 | ChEMBL |
| Ke | 0.05 nM | PMID26342544, PMID20568781, PMID19954245, PMID23651437, PMID16509593, PMID17685652 | ChEMBL |
| Ke | 0.11 nM | PMID21570305, PMID21684752, PMID17625813, PMID15743210, PMID19053757 | ChEMBL |
| Ki | 0.01 - 1.25893 nM | PMID9686407, PMID7869844, PMID7624359, PMID9262330, PMID2444704 | IUPHAR |
| Ki | 0.02 nM | PMID8114680 | BindingDB |
| Ki | 0.027 nM | PMID19027293 | ChEMBL |
| Ki | 0.027 nM | PMID19027293 | BindingDB |
| Ki | 0.039 nM | PMID11055333 | ChEMBL |
| Ki | 0.039 nM | PMID11055333 | BindingDB |
| Ki | 0.07 nM | PMID11495579 | BindingDB |
| Ki | 0.07 nM | PMID11495579 | ChEMBL |
| Ki | 0.1259 nM | PMID17490886 | ChEMBL |
| Ki | 0.13 nM | PMID10780914 | BindingDB,ChEMBL |
| Ki | 0.15 nM | PMID25513968 | ChEMBL |
| Ki | 0.15 nM | PMID25513968 | BindingDB |
| Ki | 0.153 nM | PMID21958337 | BindingDB |
| Ki | 0.153 nM | PMID21958337 | ChEMBL |
| Ki | 0.19 nM | PMID21621410 | ChEMBL |
| Ki | 0.19 nM | PMID21621410 | BindingDB |
| Ki | 0.2 nM | PMID12672258, PMID12825951, PMID12519069, PMID15456250, PMID11055333, PMID24973818, PMID14640558 | BindingDB |
| Ki | 0.2 nM | PMID12672258, PMID9686407, PMID12825951, PMID12519069, PMID15456250, PMID11055333, PMID24973818, PMID14640558 | BindingDB,ChEMBL |
| Ki | 0.23 nM | PMID21621410 | ChEMBL |
| Ki | 0.23 nM | PMID21621410 | BindingDB |
| Ki | 0.34 nM | PMID20441176 | BindingDB |
| Ki | 0.34 nM | PMID20441176 | ChEMBL |
| Ki | 0.36 nM | PMID10893307 | BindingDB |
| Ki | 0.36 nM | PMID10893307 | ChEMBL |
| Ki | 0.37 nM | PMID12747782 | BindingDB |
| Ki | 0.37 nM | PMID12747782 | ChEMBL |
| Ki | 0.4 nM | PMID15743210 | ChEMBL |
| Ki | 0.85 nM | PMID21744827 | BindingDB |
| Ki | 0.85 nM | PMID21744827 | ChEMBL |
| Ki | 0.9 nM | PMID20727752 | ChEMBL |
| Ki | 0.9 nM | PMID20727752 | BindingDB |
| Ki | 12.8 nM | PMID10780914 | BindingDB,ChEMBL |
| Ki | 12.9 nM | PMID10780914 | BindingDB,ChEMBL |
| Max inhibition | 100.0 % | PMID12747782 | ChEMBL |
| Max stimulation | -13.0 % | PMID12747782 | ChEMBL |
| Ratio | 0.01 - | PMID12565965 | ChEMBL |
| Ratio | 181.0 - | PMID1849995 | ChEMBL |
| Ratio | 473.0 - | PMID1849995 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417