

You can:
| Name | Muscarinic acetylcholine receptor M1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM1 |
| Synonym | cholinergic receptor cholinergic receptor, muscarinic 1 cholinergic receptor, muscarinic 1, CNS Chrm-1 M1 receptor [ Show all ] |
| Disease | Functional bowel syndrome; Irritable bowel syndrome Glaucoma Peptic ulcer Parkinsonism; Extrapyramidal disorders secondary to neuroleptic drug therapy Visceral spasms [ Show all ] |
| Length | 460 |
| Amino acid sequence | MNTSAPPAVSPNITVLAPGKGPWQVAFIGITTGLLSLATVTGNLLVLISFKVNTELKTVNNYFLLSLACADLIIGTFSMNLYTTYLLMGHWALGTLACDLWLALDYVASNASVMNLLLISFDRYFSVTRPLSYRAKRTPRRAALMIGLAWLVSFVLWAPAILFWQYLVGERTVLAGQCYIQFLSQPIITFGTAMAAFYLPVTVMCTLYWRIYRETENRARELAALQGSETPGKGGGSSSSSERSQPGAEGSPETPPGRCCRCCRAPRLLQAYSWKEEEEEDEGSMESLTSSEGEEPGSEVVIKMPMVDPEAQAPTKQPPRSSPNTVKRPTKKGRDRAGKGQKPRGKEQLAKRKTFSLVKEKKAARTLSAILLAFILTWTPYNIMVLVSTFCKDCVPETLWELGYWLCYVNSTINPMCYALCNKAFRDTFRLLLLCRWDKRRWRKIPKRPGSVHRTPSRQC |
| UniProt | P11229 |
| Protein Data Bank | 5cxv |
| GPCR-HGmod model | P11229 |
| 3D structure model | This structure is from PDB ID 5cxv. |
| BioLiP | BL0339262, BL0339261, BL0339263 |
| Therapeutic Target Database | T28893 |
| ChEMBL | CHEMBL216 |
| IUPHAR | 13 |
| DrugBank | BE0000092 |
| Name | CHEMBL213709 |
|---|---|
| Molecular formula | C39H45F3N4O4 |
| IUPAC name | (2R)-N-[(1-ethylpiperidin-4-yl)methyl]-1-[(2S,4R)-4-hydroxy-1-[3,3,3-tris(4-fluorophenyl)propanoyl]pyrrolidine-2-carbonyl]pyrrolidine-2-carboxamide |
| Molecular weight | 690.808 |
| Hydrogen bond acceptor | 8 |
| Hydrogen bond donor | 2 |
| XlogP | 5.1 |
| Synonyms | BDBM50194657 SCHEMBL4611569 (2R)-N-(1-ethyl-4-piperidinylmethyl)-1-((2S,4R)-4-hydroxy-1-[3,3,3-tris(4-fluorophenyl)propanoyl]pyrrolidine-2-yl)carbonylpyrrolidine-2-carboxamide |
| Inchi Key | AOYBULVQHCLRGS-VATRIRCNSA-N |
| Inchi ID | InChI=1S/C39H45F3N4O4/c1-2-44-20-17-26(18-21-44)24-43-37(49)34-4-3-19-45(34)38(50)35-22-33(47)25-46(35)36(48)23-39(27-5-11-30(40)12-6-27,28-7-13-31(41)14-8-28)29-9-15-32(42)16-10-29/h5-16,26,33-35,47H,2-4,17-25H2,1H3,(H,43,49)/t33-,34-,35+/m1/s1 |
| PubChem CID | 16086061 |
| ChEMBL | CHEMBL213709 |
| IUPHAR | N/A |
| BindingDB | 50194657 |
| DrugBank | N/A |
Structure | 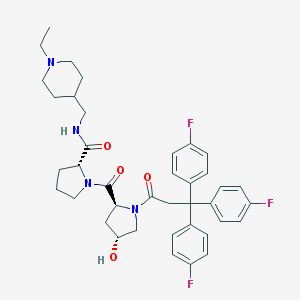 |
| Lipinski's druglikeness | This ligand is heavier than 500 daltons. This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | 580.0 nM | PMID16970392 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417