

You can:
| Name | D(2) dopamine receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Drd2 |
| Synonym | D2 receptor D2(415) and D2(444) D2A and D2B D2R Dopamine D2 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 444 |
| Amino acid sequence | MDPLNLSWYDDDLERQNWSRPFNGSEGKADRPHYNYYAMLLTLLIFIIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMIAIVWVLSFTISCPLLFGLNNTDQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRKRRKRVNTKRSSRAFRANLKTPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRMDAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSNPDSPAKPEKNGHAKIVNPRIAKFFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFMKILHC |
| UniProt | P61169 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL339 |
| IUPHAR | 215 |
| DrugBank | N/A |
| Name | UNII-UGT5535REQ |
|---|---|
| Molecular formula | C17H18ClNO |
| IUPAC name | (5R)-8-chloro-3-methyl-5-phenyl-1,2,4,5-tetrahydro-3-benzazepin-7-ol |
| Molecular weight | 287.787 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 1 |
| XlogP | 4.0 |
| Synonyms | PDSP1_000615 SCH 23390,R(+) 1H-3-Benzazepin-7-ol, 2,3,4,5-tetrahydro-8-chloro-3-methyl-5-phenyl-, (R)- Biomol-NT_000016 CHEMBL62 [ Show all ] |
| Inchi Key | GOTMKOSCLKVOGG-OAHLLOKOSA-N |
| Inchi ID | InChI=1S/C17H18ClNO/c1-19-8-7-13-9-16(18)17(20)10-14(13)15(11-19)12-5-3-2-4-6-12/h2-6,9-10,15,20H,7-8,11H2,1H3/t15-/m1/s1 |
| PubChem CID | 3036864 |
| ChEMBL | CHEMBL62 |
| IUPHAR | N/A |
| BindingDB | 82247 |
| DrugBank | N/A |
Structure | 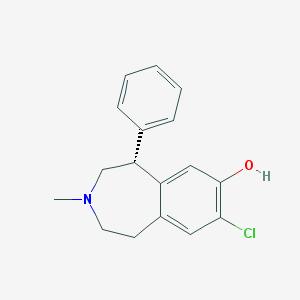 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | <10000.0 nM | PMID1971308, PMID2405158 | BindingDB,ChEMBL |
| IC50 | 600.0 nM | PMID3172140 | BindingDB,ChEMBL |
| IC50 | 1900.0 nM | PMID3172140 | BindingDB,ChEMBL |
| Inhibition | 53.0 % | PMID3039136 | ChEMBL |
| K0.5 | <500.0 nM | PMID7636869 | ChEMBL |
| K0.5 | 600.0 nM | PMID8568818 | ChEMBL |
| Ki | <1000.0 nM | PMID8863801 | BindingDB,ChEMBL |
| Ki | 0.537 nM | PMID10425088 | ChEMBL |
| Ki | 359.0 nM | PMID2905002 | BindingDB |
| Ki | 569.0 nM | PMID11044891 | BindingDB |
| Ki | 600.0 nM | PMID2966245 | BindingDB,ChEMBL |
| Ki | 648.0 nM | PMID2666667 | BindingDB |
| Ki | 648.0 nM | PMID2666667 | ChEMBL |
| Ki | 676.08 nM | PMID7996543 | ChEMBL |
| Ki | 760.0 nM | , PMID6387355, Bioorg. Med. Chem. Lett., (1992) 2:5:399 | BindingDB,ChEMBL |
| Ki | 820.0 nM | PMID2136916 | BindingDB,ChEMBL |
| Ki | 886.0 nM | PMID1833546 | BindingDB,ChEMBL |
| Ki | 895.0 nM | PMID2415793, PMID1397049, PMID7855175 | BindingDB |
| Ki | 900.0 nM | PMID3050089, PMID1397049 | BindingDB,ChEMBL |
| Ki | 2367.0 nM | PMID1356154 | BindingDB |
| Ki | 2500.0 nM | PMID2532362 | BindingDB |
| Ki | 3300.0 nM | PMID2532362 | BindingDB |
| Ki | 4650.0 nM | PMID8278895 | BindingDB |
| Ki | 5000.0 nM | PMID12023552 | BindingDB |
| Ki | 5560.0 nM | PMID1356154 | BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417