Back to FUpred homepage
About FUpred Server
Overview
The pipeline of the FUpred algorithm is shown Figure 1.
Starting from the input protein sequence,
a deep MSA is generated by iterative sequence homolog searches against multiple sequence databases.
Then, with the deep MSA as input, the secondary structure of a query sequence is predicted by PSSpred,
and the contact-map is predicted with the ResPRE method.
Both the contact-map and secondary structure information are used in FU-score calculation and revision based on a recursion strategy to detect domain boundary.
The recursion strategy is based on the difference between the distributions of multi- and single-domain.
In detail, with the contact-map and secondary structure information of the query sequence as input,
both FU-score2c and FU-score2d are calculated for the input sequence.
Due to the differences between distributions of multi- and single-domain proteins,
we can use two cutoffs, Cutoff2c and Cutoff2d,
to distinguish continuous multi- and single-domain proteins,
as well as discontinuous multi- and single-domain proteins, respectively.
If the FU-score2c/FU-score2d of the input protein is less than the Cutoff2c/Cutoff2d,
the input protein is predicted as continuous/discontinuous two-domain protein with the domain boundaries generated by FU-score;
otherwise, the input protein is predicted as single domain protein.
When both the FU-score2c and FU-score2d of the input protein are less than the Cutoff2c and Cutoff2d,
respectively, we compare the differences (Cutoff2c minus FU-score2c) and (Cutoff2d minus FU-score2d),
and adopt the domain boundaries predicted by the method with larger difference.
Then, the two possible domains recursively perform the same procedure as shown above.
The recursion procedure is stopped when none of domains can be further split.
FUpred flowchart
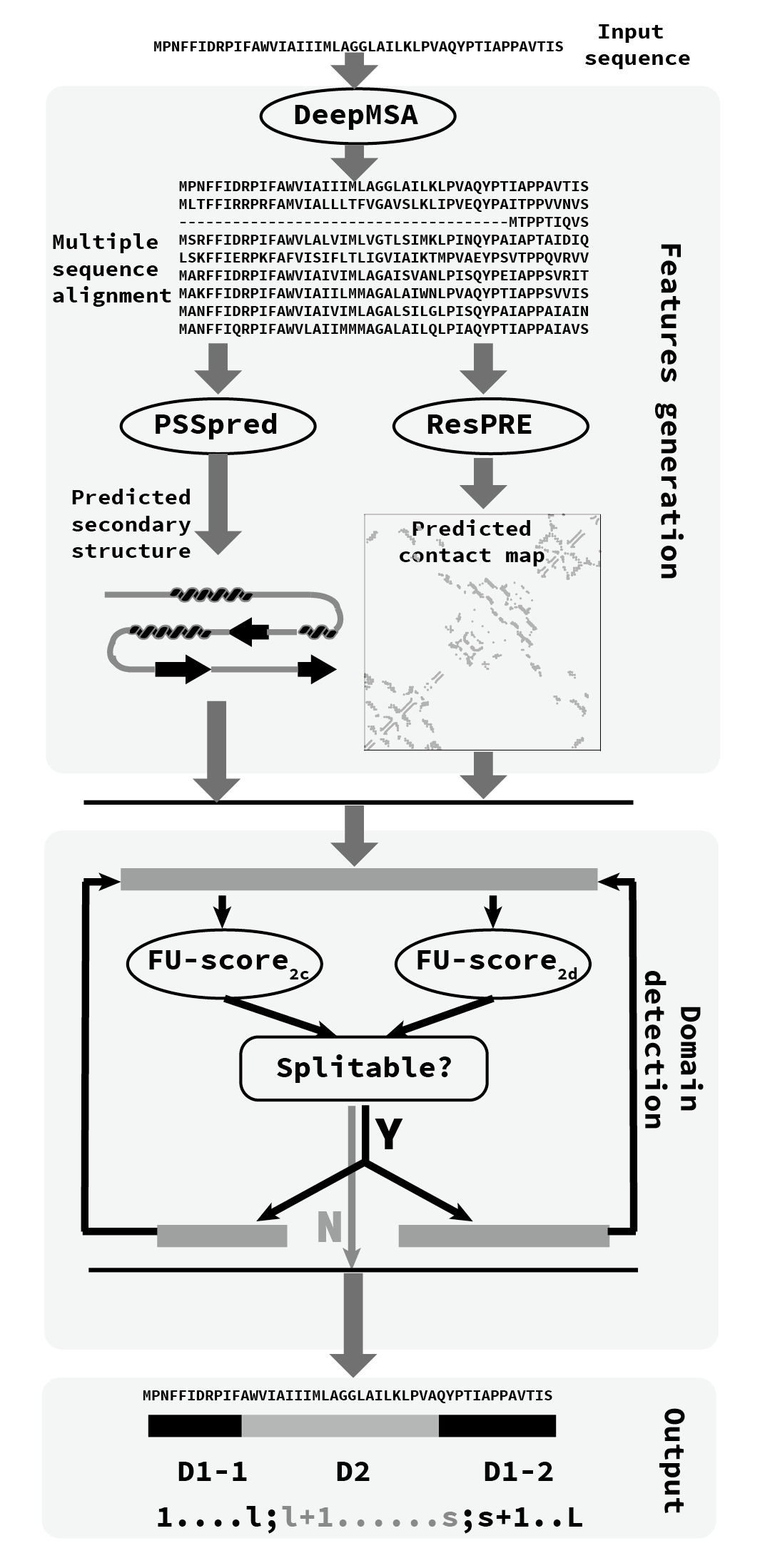
Figure 1. Pipeline of FUpred.
User Inputs
The user need to paste the FASTA format amino acid sequence to input box, or upload the amino acid sequence of the query protein through browser button.
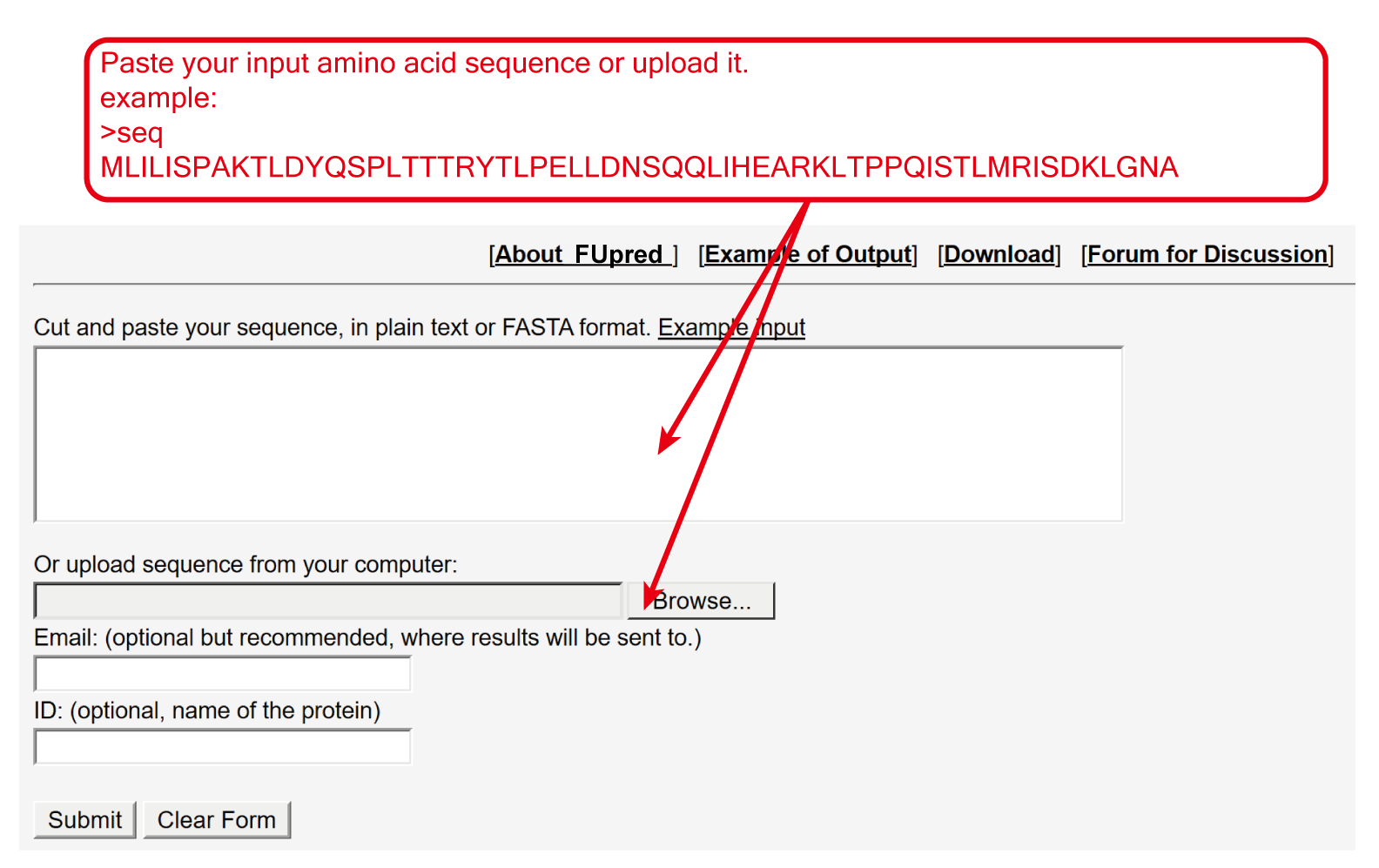
Figure 2. User inputs.
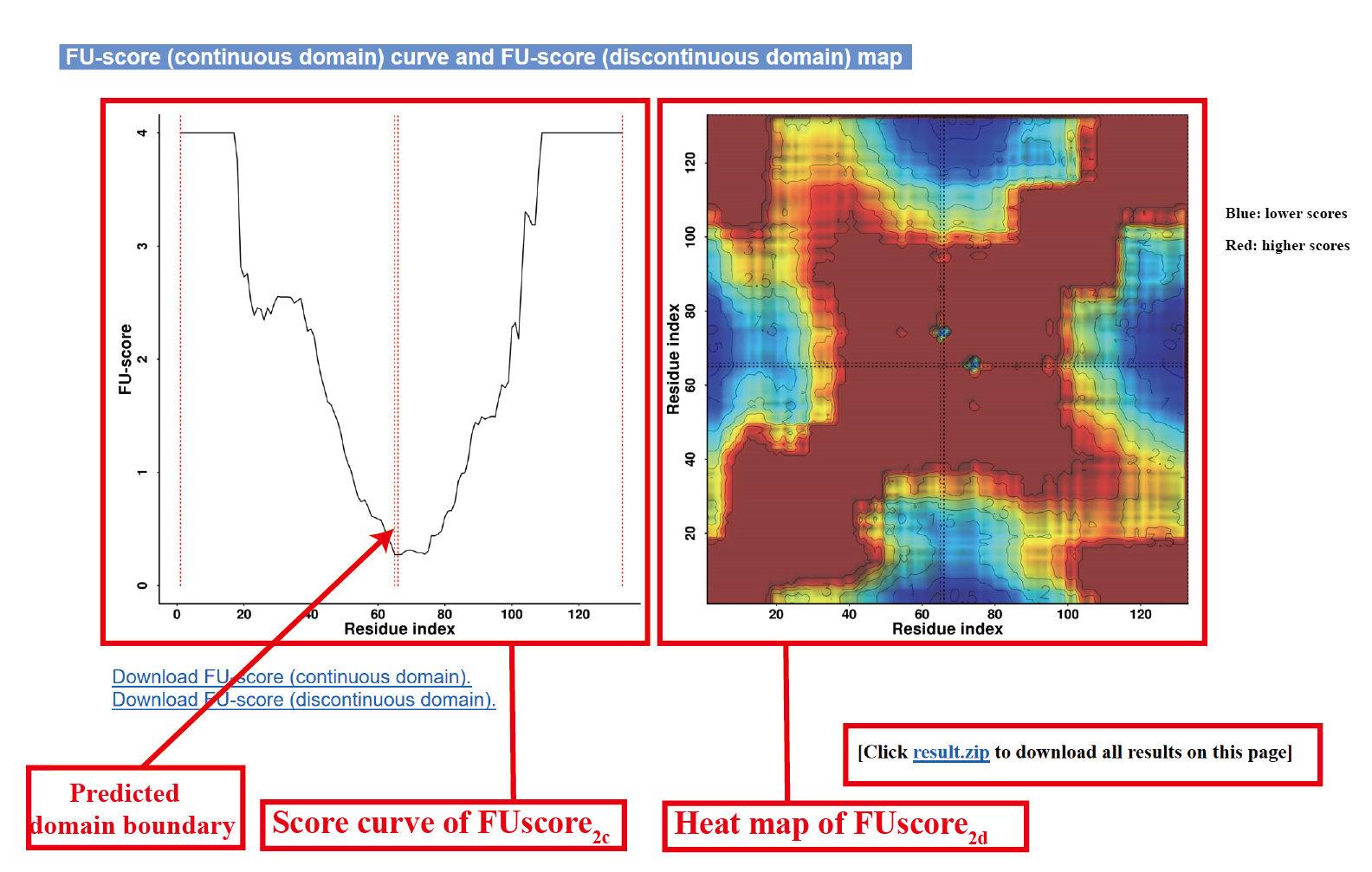
Content in output page
The outputs of FUpred include:
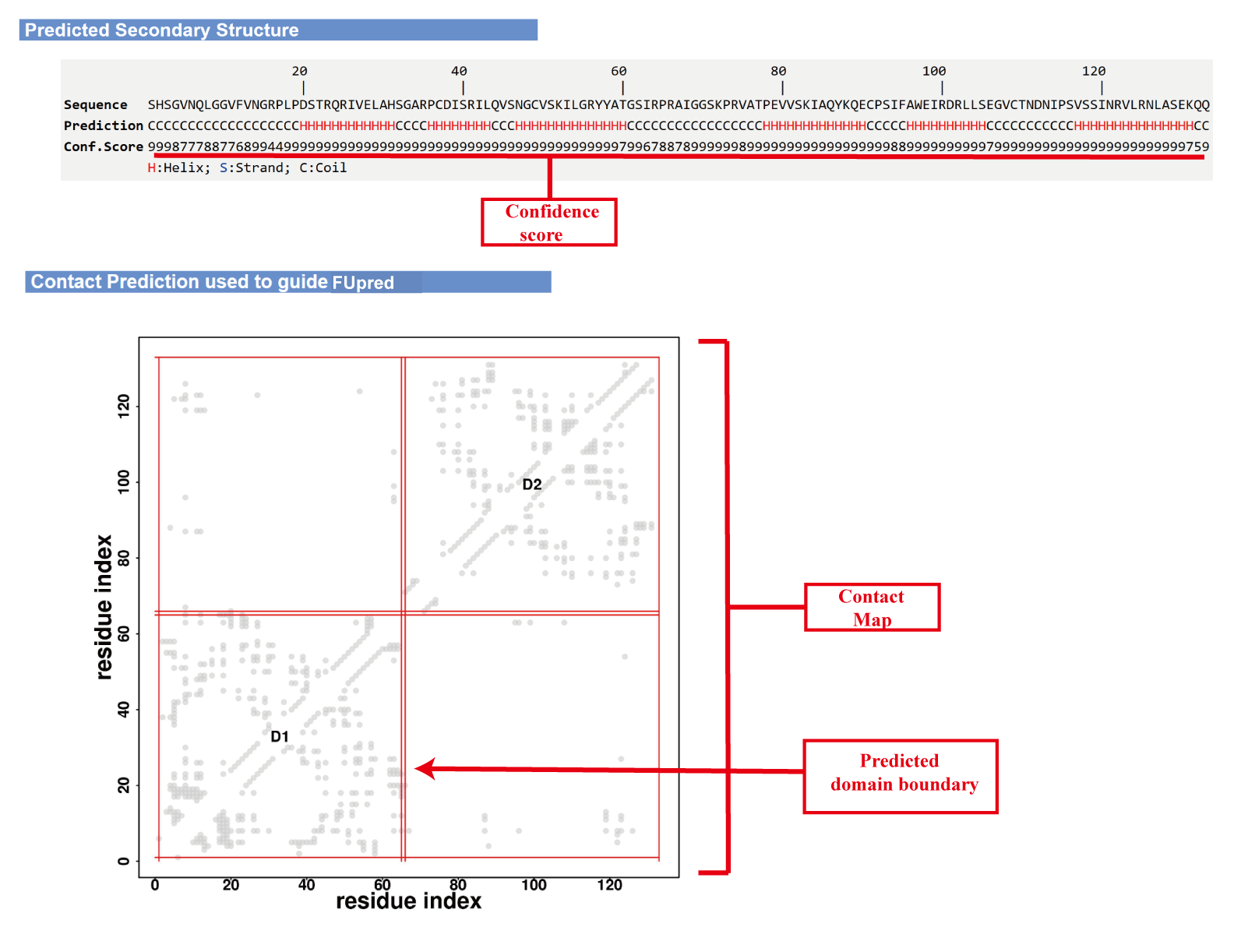
(i) predicted secondary structure and contact map;
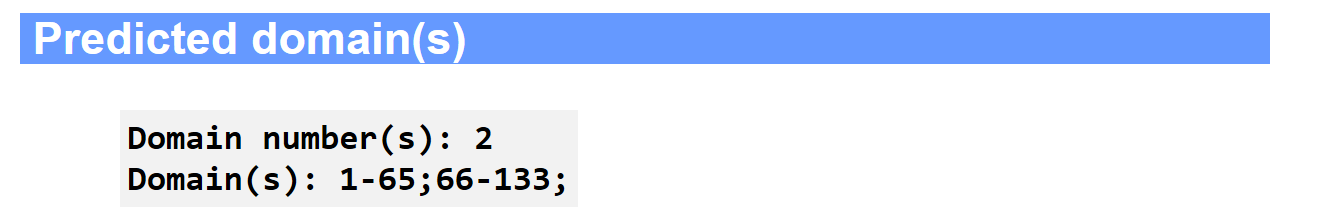
(ii) predicted domain boundary;
(iii) FU-score curve and FU-score map;
Illustration of outputs
Standalone package instruction
References:
Wei Zheng, Xiaogen Zhou, Qiqige Wuyun, Robin Pearce, Yang Li and Yang Zhang.
FUpred: Detecting protein domains through deep-learning based contact map prediction.
in press, Bioinformatics (2020).
Back to the FUpred homepage




![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417