

You can:
| Name | Substance-K receptor |
|---|---|
| Species | Oryctolagus cuniculus (Rabbit) |
| Gene | TACR2 |
| Synonym | Neurokinin A receptor NK-2 receptor NK-2R SKR Tachykinin receptor 2 |
| Disease | N/A for non-human GPCRs |
| Length | 384 |
| Amino acid sequence | MGACDIVTEANISSDIDSNATGVTAFSMPGWQLALWATAYLALVLVAVVGNATVIWIILAHRRMRTVTNYFIVNLALADLCMATFNAAFNFVYASHNIWYFGRAFCYFQNLFPITAMFVSIYSMTAIAADRYMAIVHPFQPRLSGPGTKAVIAGIWLVALALAFPQCFYSTITMDQGATKCVVAWPEDSGGKMLLLYHLTVIALIYFLPLVVMFVAYSVIGFKLWRRTVPGHQTHGANLRHLRAKKKFVKTMVLVVVTFAVCWLPYHLYFLLGHFQDDIYCRKFIQQVYLVLFWLAMSSTMYNPIIYCCLNHRFRSGFRLAFRCCPWVTPTEEDKLELTHTPSLSVRVNRCHTKETLFLVGDVAPSEAANGQAGGPQDGGAYDF |
| UniProt | P79218 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3433 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 776 | 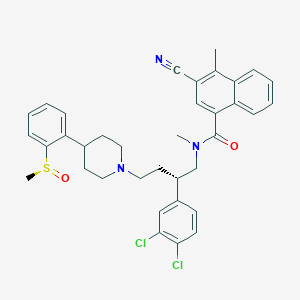 CHEMBL408242 CHEMBL408242 | C36H37Cl2N3O2S | 646.671 | 5 / 0 | 7.5 | No |
| 1721 | 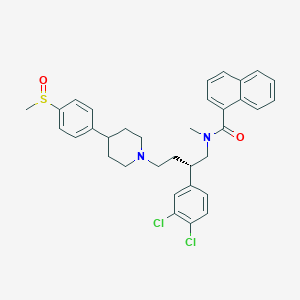 CHEMBL2373196 CHEMBL2373196 | C34H36Cl2N2O2S | 607.634 | 4 / 0 | 7.4 | No |
| 8429 | 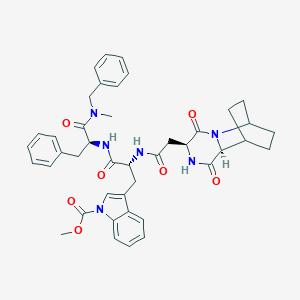 CHEMBL3144347 CHEMBL3144347 | C42H46N6O7 | 746.865 | 7 / 3 | 4.2 | No |
| 519733 | 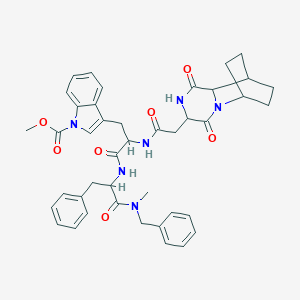 CHEMBL45085 CHEMBL45085 | C42H46N6O7 | 746.865 | 7 / 3 | 4.2 | No |
| 12965 | 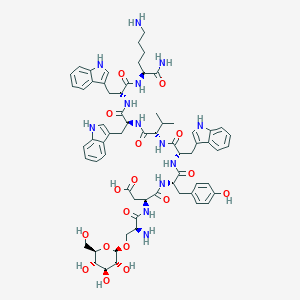 CHEMBL2370585 CHEMBL2370585 | C66H83N13O17 | 1330.46 | 19 / 19 | -1.8 | No |
| 13058 | 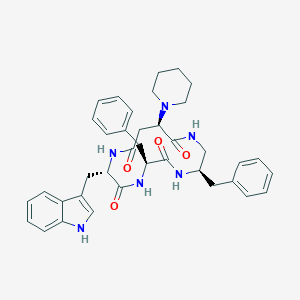 CHEMBL387699 CHEMBL387699 | C38H44N6O4 | 648.808 | 5 / 5 | 4.3 | No |
| 18001 | 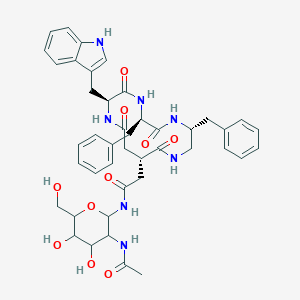 CHEMBL161288 CHEMBL161288 | C43H51N7O10 | 825.92 | 10 / 10 | 0.5 | No |
| 18003 | 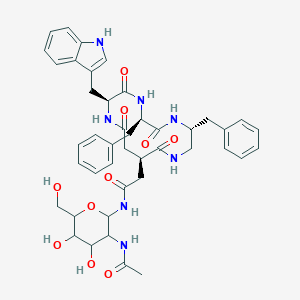 CHEMBL350596 CHEMBL350596 | C43H51N7O10 | 825.92 | 10 / 10 | 0.5 | No |
| 519780 | 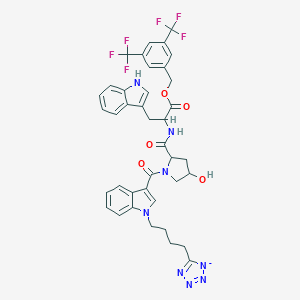 CHEMBL65468 CHEMBL65468 | C39H35F6N8O5- | 809.75 | 15 / 3 | 5.2 | No |
| 20128 | 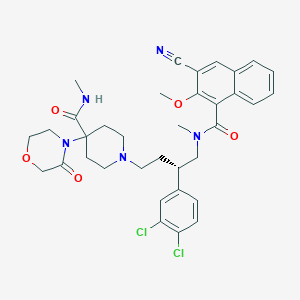 CHEMBL337435 CHEMBL337435 | C35H39Cl2N5O5 | 680.627 | 7 / 1 | 4.6 | No |
| 20472 | 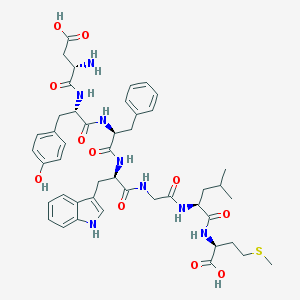 CHEMBL329888 CHEMBL329888 | C46H58N8O11S | 931.075 | 13 / 11 | 0.6 | No |
| 24202 | 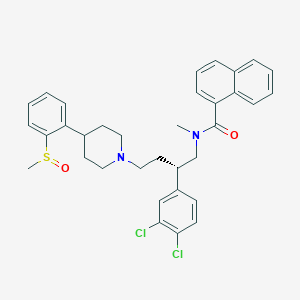 CHEMBL2373189 CHEMBL2373189 | C34H36Cl2N2O2S | 607.634 | 4 / 0 | 7.4 | No |
| 28386 | 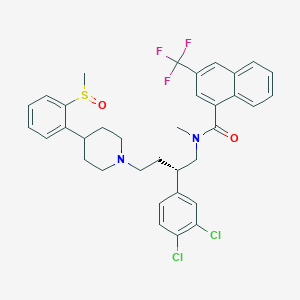 CHEMBL96794 CHEMBL96794 | C35H35Cl2F3N2O2S | 675.632 | 7 / 0 | 8.3 | No |
| 30745 | 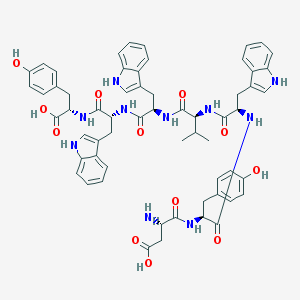 CHEMBL412621 CHEMBL412621 | C60H64N10O12 | 1117.23 | 13 / 14 | 3.2 | No |
| 31970 | 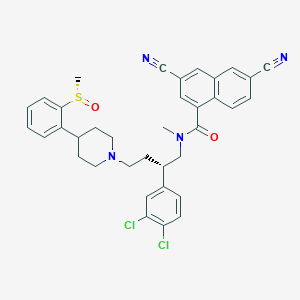 CHEMBL128723 CHEMBL128723 | C36H34Cl2N4O2S | 657.654 | 6 / 0 | 6.8 | No |
| 33182 | 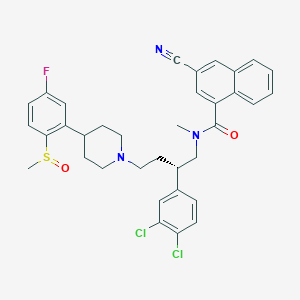 CHEMBL341016 CHEMBL341016 | C35H34Cl2FN3O2S | 650.634 | 6 / 0 | 7.2 | No |
| 36213 | 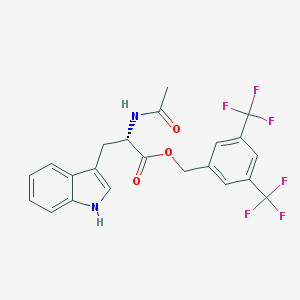 L-732,138 L-732,138 | C22H18F6N2O3 | 472.387 | 9 / 2 | 4.7 | Yes |
| 37513 | 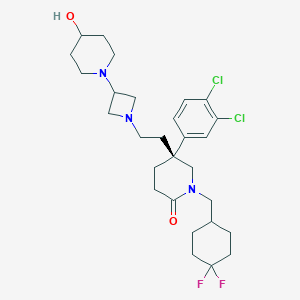 CHEMBL434548 CHEMBL434548 | C28H39Cl2F2N3O2 | 558.536 | 6 / 1 | 5.1 | No |
| 46026 | 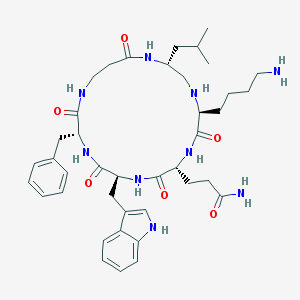 CHEMBL332820 CHEMBL332820 | C40H57N9O6 | 759.953 | 8 / 9 | 1.5 | No |
| 46180 | 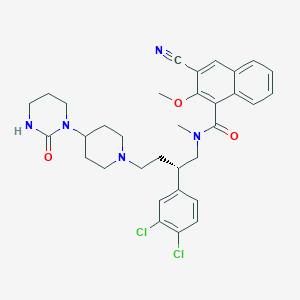 CHEMBL128782 CHEMBL128782 | C33H37Cl2N5O3 | 622.591 | 5 / 1 | 5.7 | No |
| 49585 | 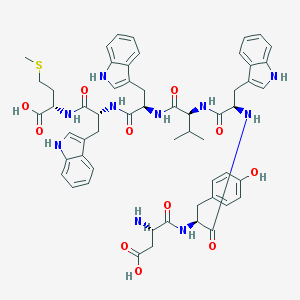 CHEMBL387199 CHEMBL387199 | C56H64N10O11S | 1085.25 | 13 / 13 | 2.6 | No |
| 49799 | 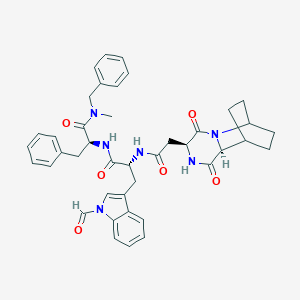 CHEMBL3144343 CHEMBL3144343 | C41H44N6O6 | 716.839 | 6 / 3 | 3.8 | No |
| 519911 | 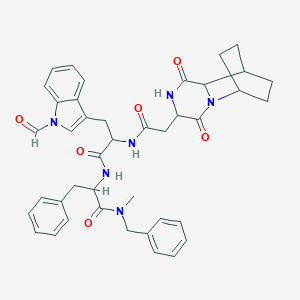 CHEMBL296267 CHEMBL296267 | C41H44N6O6 | 716.839 | 6 / 3 | 3.8 | No |
| 51868 | 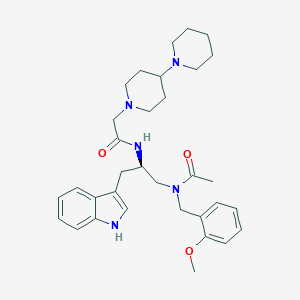 LANEPITANT LANEPITANT | C33H45N5O3 | 559.755 | 5 / 2 | 4.2 | No |
| 51871 | 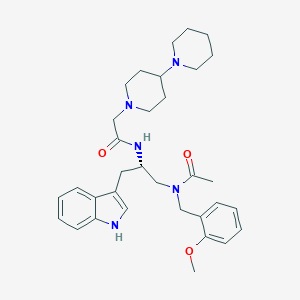 CHEMBL351236 CHEMBL351236 | C33H45N5O3 | 559.755 | 5 / 2 | 4.2 | No |
| 53511 | 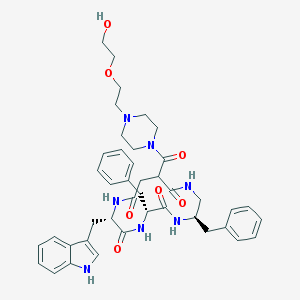 CHEMBL161242 CHEMBL161242 | C42H51N7O7 | 765.912 | 8 / 6 | 2.0 | No |
| 53742 | 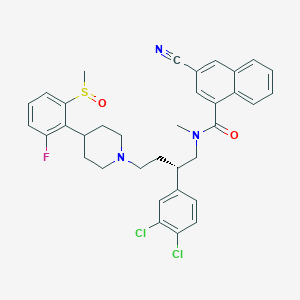 CHEMBL132347 CHEMBL132347 | C35H34Cl2FN3O2S | 650.634 | 6 / 0 | 7.2 | No |
| 55332 | 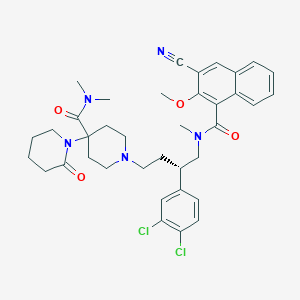 CHEMBL128891 CHEMBL128891 | C37H43Cl2N5O4 | 692.682 | 6 / 0 | 5.5 | No |
| 55348 | 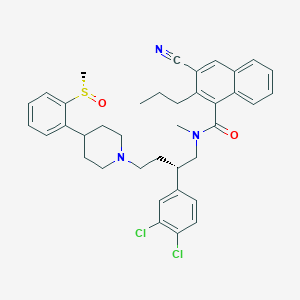 CHEMBL146796 CHEMBL146796 | C38H41Cl2N3O2S | 674.725 | 5 / 0 | 8.4 | No |
| 58150 | 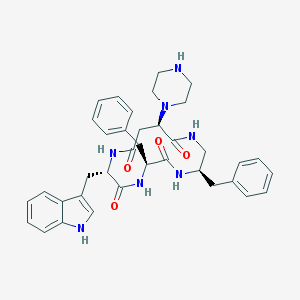 CHEMBL223644 CHEMBL223644 | C37H43N7O4 | 649.796 | 6 / 6 | 2.8 | No |
| 59415 | 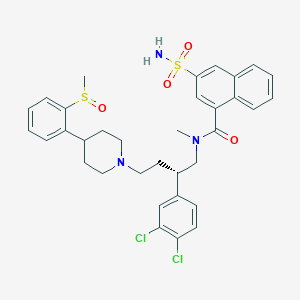 CHEMBL411037 CHEMBL411037 | C34H37Cl2N3O4S2 | 686.707 | 7 / 1 | 6.0 | No |
| 62961 | 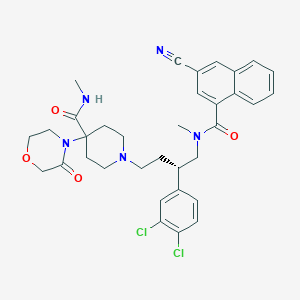 CHEMBL130085 CHEMBL130085 | C34H37Cl2N5O4 | 650.601 | 6 / 1 | 4.6 | No |
| 64537 | 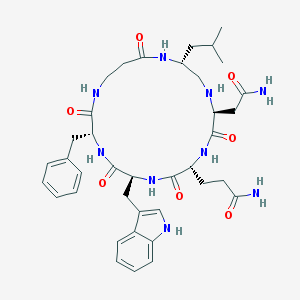 CHEMBL433192 CHEMBL433192 | C38H51N9O7 | 745.882 | 8 / 9 | 0.2 | No |
| 64676 | 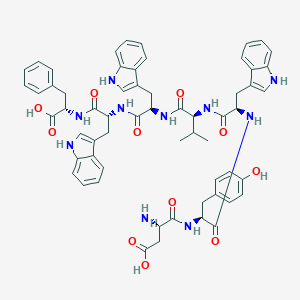 CHEMBL412189 CHEMBL412189 | C60H64N10O11 | 1101.23 | 12 / 13 | 3.5 | No |
| 65549 | 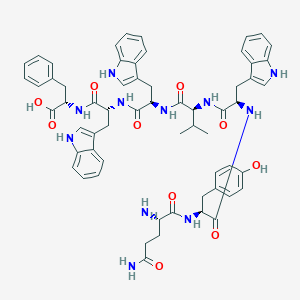 CHEMBL268563 CHEMBL268563 | C61H67N11O10 | 1114.27 | 11 / 13 | 3.3 | No |
| 68234 | 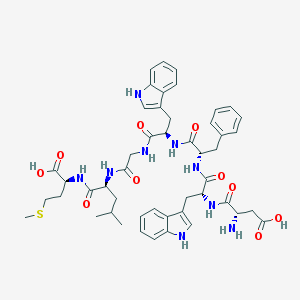 CHEMBL327457 CHEMBL327457 | C48H59N9O10S | 954.113 | 12 / 11 | 1.1 | No |
| 68569 | 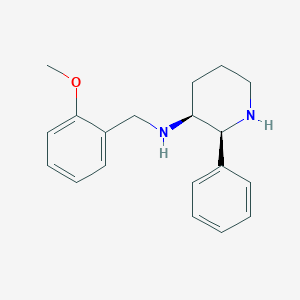 CP-99994 CP-99994 | C19H24N2O | 296.414 | 3 / 2 | 2.9 | Yes |
| 70781 | 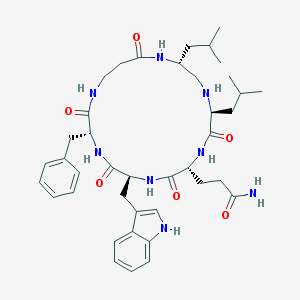 CHEMBL421062 CHEMBL421062 | C40H56N8O6 | 744.938 | 7 / 8 | 3.0 | No |
| 73432 | 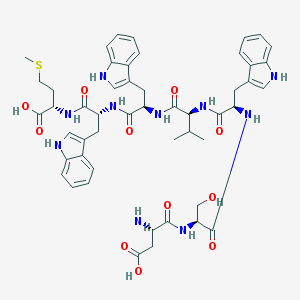 CHEMBL441181 CHEMBL441181 | C50H60N10O11S | 1009.15 | 13 / 13 | 0.3 | No |
| 80400 | 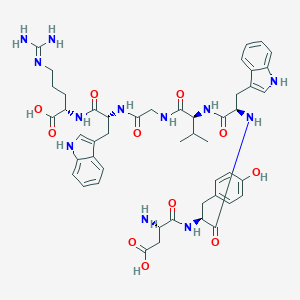 CHEMBL438716 CHEMBL438716 | C48H60N12O11 | 981.081 | 13 / 14 | -1.4 | No |
| 80404 | 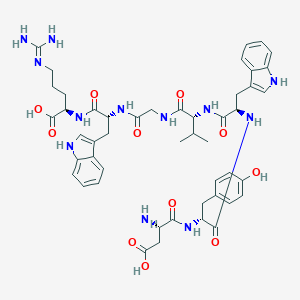 CHEMBL266526 CHEMBL266526 | C48H60N12O11 | 981.081 | 13 / 14 | -1.4 | No |
| 81433 | 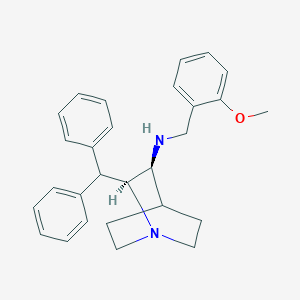 CP 96345 CP 96345 | C28H32N2O | 412.577 | 3 / 1 | 5.4 | No |
| 85463 | 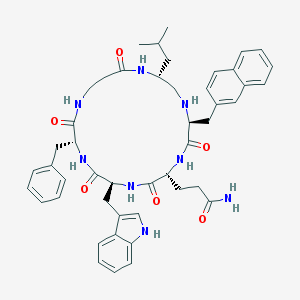 CHEMBL121271 CHEMBL121271 | C47H56N8O6 | 829.015 | 7 / 8 | 4.5 | No |
| 90388 | 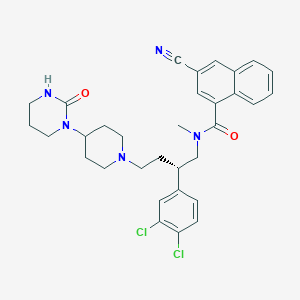 CHEMBL340736 CHEMBL340736 | C32H35Cl2N5O2 | 592.565 | 4 / 1 | 5.8 | No |
| 93122 | 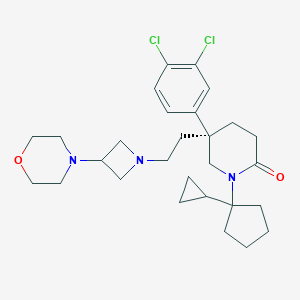 CHEMBL360134 CHEMBL360134 | C28H39Cl2N3O2 | 520.539 | 4 / 0 | 4.5 | No |
| 99479 | 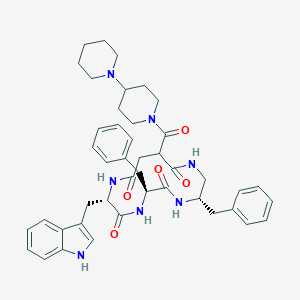 CHEMBL2112922 CHEMBL2112922 | C44H53N7O5 | 759.952 | 6 / 5 | 4.5 | No |
| 102778 | 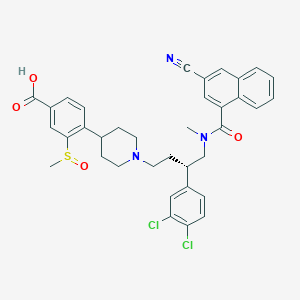 CHEMBL339575 CHEMBL339575 | C36H35Cl2N3O4S | 676.653 | 7 / 1 | 4.4 | No |
| 105592 | 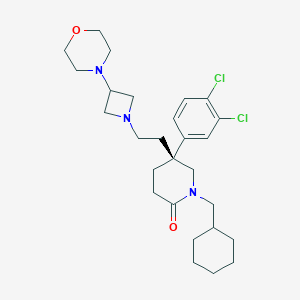 CHEMBL185572 CHEMBL185572 | C27H39Cl2N3O2 | 508.528 | 4 / 0 | 5.1 | No |
| 108439 | 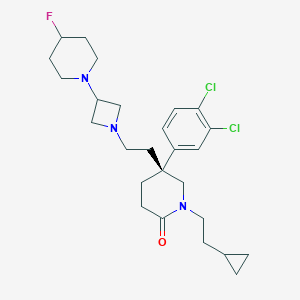 CHEMBL185133 CHEMBL185133 | C26H36Cl2FN3O | 496.492 | 4 / 0 | 5.4 | No |
| 112455 | 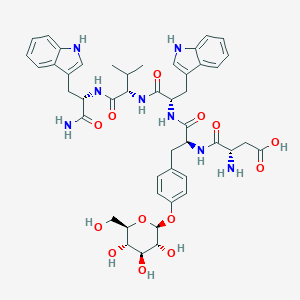 CHEMBL3215392 CHEMBL3215392 | C46H56N8O13 | 928.997 | 14 / 13 | -1.7 | No |
| 112588 | 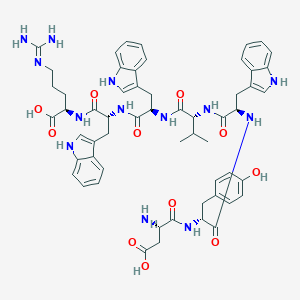 CHEMBL406487 CHEMBL406487 | C57H67N13O11 | 1110.24 | 13 / 15 | 0.8 | No |
| 112591 | 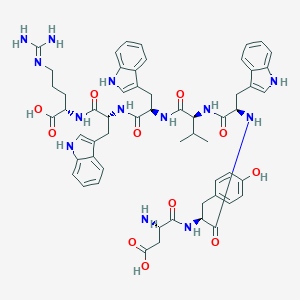 CHEMBL429340 CHEMBL429340 | C57H67N13O11 | 1110.24 | 13 / 15 | 0.8 | No |
| 112593 |  CHEMBL217365 CHEMBL217365 | C57H67N13O11 | 1110.24 | 13 / 15 | 0.8 | No |
| 113824 |  CHEMBL367815 CHEMBL367815 | C28H37Cl2F2N3O2 | 556.52 | 6 / 0 | 4.7 | No |
| 520191 |  CHEMBL297376 CHEMBL297376 | C44H48N6O8 | 788.902 | 8 / 4 | 3.2 | No |
| 117949 |  CHEMBL3144351 CHEMBL3144351 | C44H48N6O8 | 788.902 | 8 / 4 | 3.2 | No |
| 119308 | 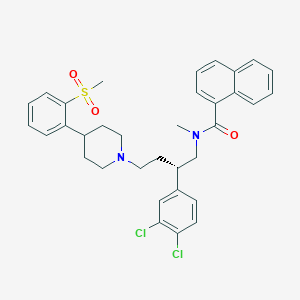 CHEMBL2373193 CHEMBL2373193 | C34H36Cl2N2O3S | 623.633 | 4 / 0 | 7.5 | No |
| 125935 | 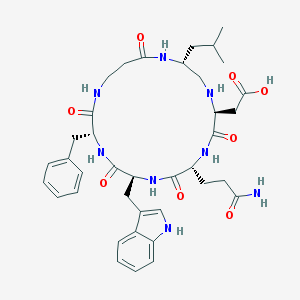 CHEMBL332872 CHEMBL332872 | C38H50N8O8 | 746.866 | 9 / 9 | -1.4 | No |
| 131792 | 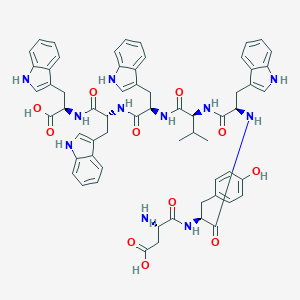 CHEMBL439287 CHEMBL439287 | C62H65N11O11 | 1140.27 | 12 / 14 | 3.7 | No |
| 138441 | 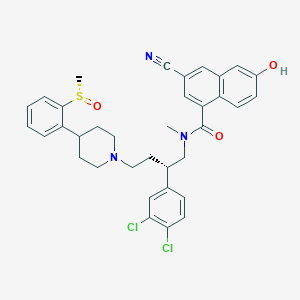 CHEMBL129117 CHEMBL129117 | C35H35Cl2N3O3S | 648.643 | 6 / 1 | 6.8 | No |
| 141353 | 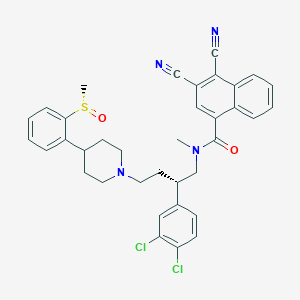 CHEMBL130082 CHEMBL130082 | C36H34Cl2N4O2S | 657.654 | 6 / 0 | 6.8 | No |
| 143625 | 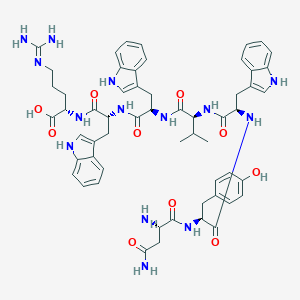 CHEMBL407328 CHEMBL407328 | C57H68N14O10 | 1109.26 | 12 / 15 | 0.1 | No |
| 150369 | 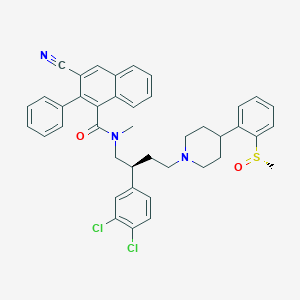 CHEMBL342758 CHEMBL342758 | C41H39Cl2N3O2S | 708.742 | 5 / 0 | 8.7 | No |
| 153733 | 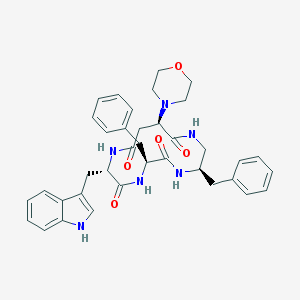 CHEMBL389459 CHEMBL389459 | C37H42N6O5 | 650.78 | 6 / 5 | 3.1 | No |
| 153736 | 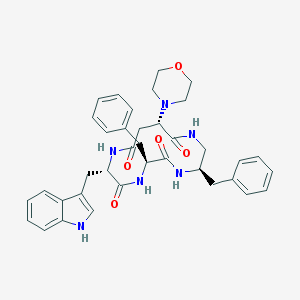 CHEMBL387698 CHEMBL387698 | C37H42N6O5 | 650.78 | 6 / 5 | 3.1 | No |
| 156147 | 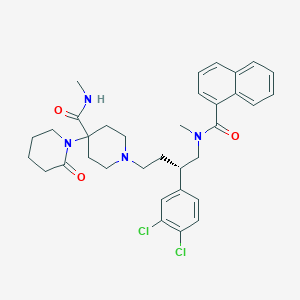 CHEMBL2373191 CHEMBL2373191 | C34H40Cl2N4O3 | 623.619 | 4 / 1 | 5.6 | No |
| 156299 | 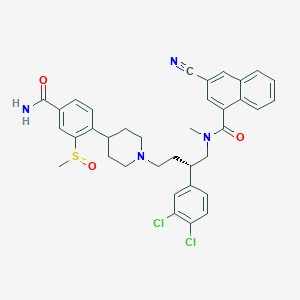 CHEMBL436388 CHEMBL436388 | C36H36Cl2N4O3S | 675.669 | 6 / 1 | 6.0 | No |
| 162910 | 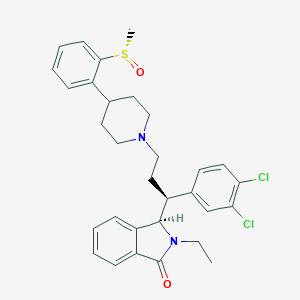 CHEMBL339767 CHEMBL339767 | C31H34Cl2N2O2S | 569.585 | 4 / 0 | 6.1 | No |
| 170404 | 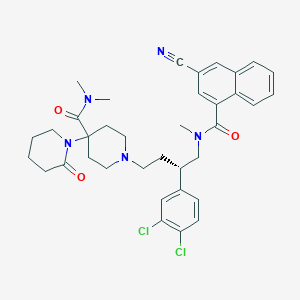 CHEMBL341175 CHEMBL341175 | C36H41Cl2N5O3 | 662.656 | 5 / 0 | 5.5 | No |
| 171324 | 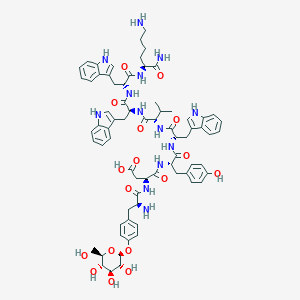 CHEMBL3215393 CHEMBL3215393 | C72H87N13O17 | 1406.56 | 19 / 19 | 0.3 | No |
| 175573 | 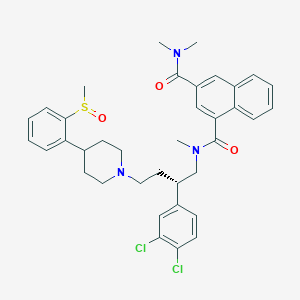 CHEMBL322879 CHEMBL322879 | C37H41Cl2N3O3S | 678.713 | 5 / 0 | 6.9 | No |
| 177875 | 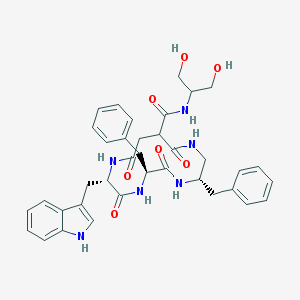 CHEMBL2112926 CHEMBL2112926 | C37H42N6O7 | 682.778 | 7 / 8 | 2.1 | No |
| 179304 |  CHEMBL333515 CHEMBL333515 | C44H62N8O6 | 799.03 | 7 / 7 | 4.9 | No |
| 180041 |  CHEMBL434142 CHEMBL434142 | C28H40Cl2F2N4O | 557.552 | 6 / 1 | 4.8 | No |
| 181179 |  CHEMBL267409 CHEMBL267409 | C53H58N10O11 | 1011.11 | 12 / 13 | 1.6 | No |
| 182790 |  CHEMBL2373197 CHEMBL2373197 | C31H36Cl2N4O2 | 567.555 | 3 / 1 | 6.0 | No |
| 183845 |  CHEMBL263743 CHEMBL263743 | C51H64N12O11 | 1021.15 | 13 / 13 | -0.7 | No |
| 184789 |  CHEMBL263985 CHEMBL263985 | C59H68N14O10 | 1133.28 | 12 / 15 | 1.2 | No |
| 184793 |  CHEMBL216897 CHEMBL216897 | C59H68N14O10 | 1133.28 | 12 / 15 | 1.2 | No |
| 185049 |  CHEMBL180049 CHEMBL180049 | C27H39Cl2N3O2 | 508.528 | 4 / 0 | 4.6 | No |
| 188044 | 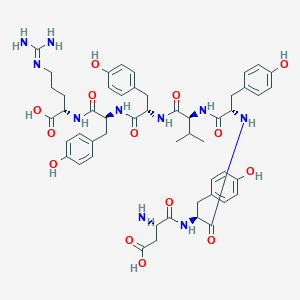 CHEMBL413730 CHEMBL413730 | C51H64N10O14 | 1041.13 | 16 / 15 | -0.7 | No |
| 188517 | 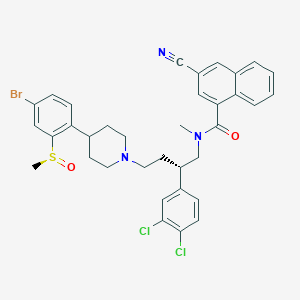 CHEMBL339213 CHEMBL339213 | C35H34BrCl2N3O2S | 711.54 | 5 / 0 | 7.8 | No |
| 520465 | 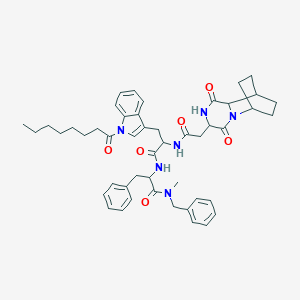 CHEMBL437060 CHEMBL437060 | C48H58N6O6 | 815.028 | 6 / 3 | 6.8 | No |
| 191046 | 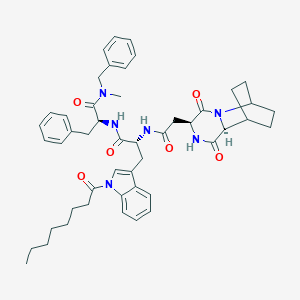 CHEMBL3144339 CHEMBL3144339 | C48H58N6O6 | 815.028 | 6 / 3 | 6.8 | No |
| 199388 |  CHEMBL121571 CHEMBL121571 | C39H54N8O6S | 762.971 | 8 / 8 | 2.3 | No |
| 200741 |  CHEMBL2112924 CHEMBL2112924 | C41H48N6O10 | 784.867 | 10 / 10 | 0.5 | No |
| 205036 |  CHEMBL3215394 CHEMBL3215394 | C63H78N12O15 | 1243.39 | 17 / 17 | -0.7 | No |
| 207152 |  CHEMBL129264 CHEMBL129264 | C36H37Cl2N3O3S2 | 694.73 | 7 / 0 | 6.2 | No |
| 207618 | 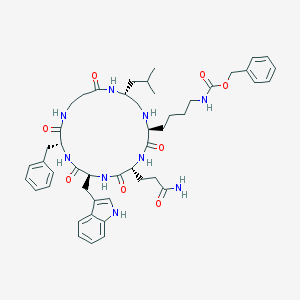 CHEMBL333027 CHEMBL333027 | C48H63N9O8 | 894.087 | 9 / 9 | 3.6 | No |
| 208371 | 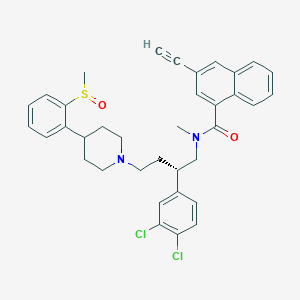 CHEMBL318364 CHEMBL318364 | C36H36Cl2N2O2S | 631.656 | 4 / 0 | 7.6 | No |
| 212824 | 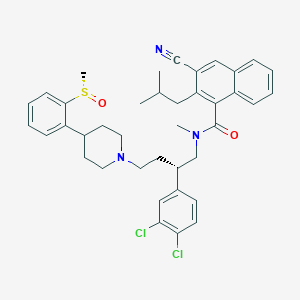 CHEMBL357908 CHEMBL357908 | C39H43Cl2N3O2S | 688.752 | 5 / 0 | 8.7 | No |
| 214332 | 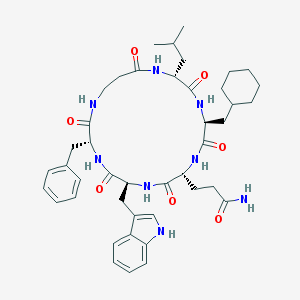 CHEMBL421071 CHEMBL421071 | C43H58N8O7 | 798.986 | 7 / 8 | 4.2 | No |
| 214444 | 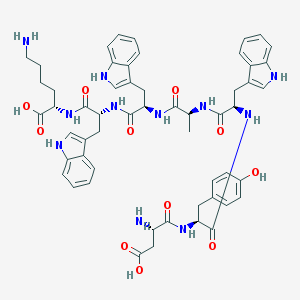 CHEMBL385914 CHEMBL385914 | C55H63N11O11 | 1054.18 | 13 / 14 | -1.5 | No |
| 218653 | 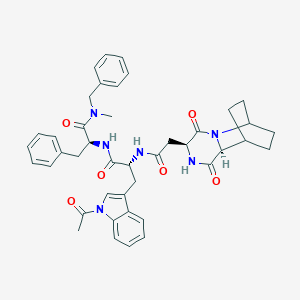 CHEMBL3144340 CHEMBL3144340 | C42H46N6O6 | 730.866 | 6 / 3 | 3.8 | No |
| 520573 | 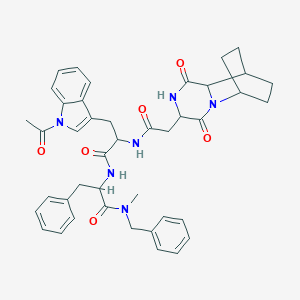 CHEMBL46849 CHEMBL46849 | C42H46N6O6 | 730.866 | 6 / 3 | 3.8 | No |
| 218708 | 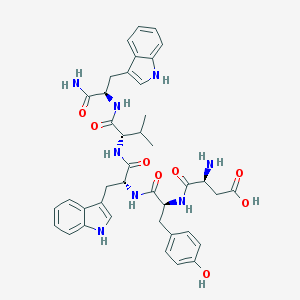 MEN-10414 MEN-10414 | C40H46N8O8 | 766.856 | 9 / 10 | -0.4 | No |
| 222288 | 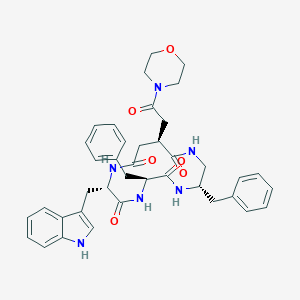 CHEMBL2112923 CHEMBL2112923 | C39H44N6O6 | 692.817 | 6 / 5 | 2.4 | No |
| 230677 | 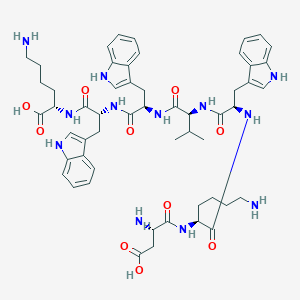 CHEMBL262647 CHEMBL262647 | C54H70N12O10 | 1047.23 | 13 / 14 | -2.0 | No |
| 231063 | 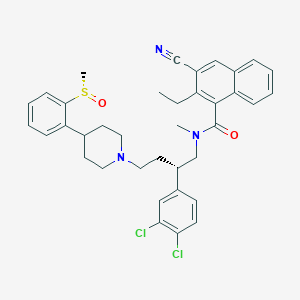 CHEMBL149326 CHEMBL149326 | C37H39Cl2N3O2S | 660.698 | 5 / 0 | 7.9 | No |
| 232425 | 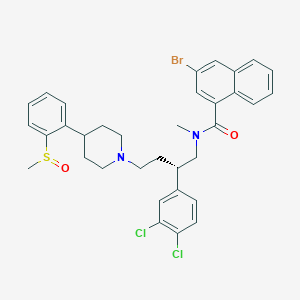 CHEMBL92742 CHEMBL92742 | C34H35BrCl2N2O2S | 686.53 | 4 / 0 | 8.1 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417