

You can:
| Name | Cannabinoid receptor 1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Cnr1 |
| Synonym | SKR6R Neuronal cannabinoid receptor Central cannabinoid receptor CB1R CB1 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 473 |
| Amino acid sequence | MKSILDGLADTTFRTITTDLLYVGSNDIQYEDIKGDMASKLGYFPQKFPLTSFRGSPFQEKMTAGDNSPLVPAGDTTNITEFYNKSLSSFKENEENIQCGENFMDMECFMILNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCRPSYHFIGSLAVADLLGSVIFVYSFVDFHVFHRKDSPNVFLFKLGGVTASFTASVGSLFLTAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCKKLQSVCSDIFPLIDETYLMFWIGVTSVLLLFIVYAYMYILWKAHSHAVRMIQRGTQKSIIIHTSEDGKVQVTRPDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMFPSCEGTAQPLDNSMGDSDCLHKHANNTASMHRAAESCIKSTVKIAKVTMSVSTDTSAEAL |
| UniProt | P20272 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3571 |
| IUPHAR | 56 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 38 | 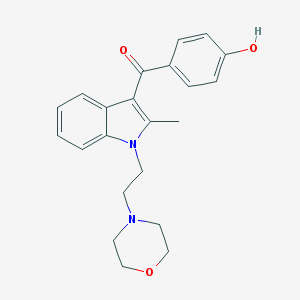 CHEMBL12859 CHEMBL12859 | C22H24N2O3 | 364.445 | 4 / 1 | 3.0 | Yes |
| 304 | 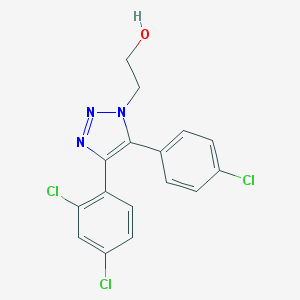 CHEMBL2237755 CHEMBL2237755 | C16H12Cl3N3O | 368.642 | 3 / 1 | 4.1 | Yes |
| 834 | 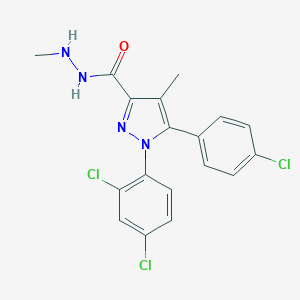 CHEMBL88548 CHEMBL88548 | C18H15Cl3N4O | 409.695 | 3 / 2 | 5.2 | No |
| 940 | 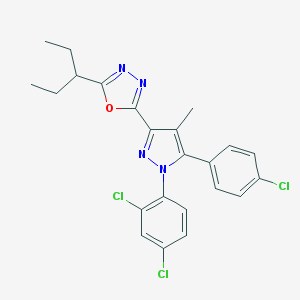 CHEMBL486958 CHEMBL486958 | C23H21Cl3N4O | 475.798 | 4 / 0 | 7.2 | No |
| 1096 |  CHEMBL231464 CHEMBL231464 | C29H44O2 | 424.669 | 2 / 1 | 9.6 | No |
| 1173 |  CHEMBL2064052 CHEMBL2064052 | C22H24O4 | 352.43 | 4 / 2 | 3.8 | Yes |
| 1451 |  CHEMBL70952 CHEMBL70952 | C25H24N2O | 368.48 | 2 / 1 | 5.0 | Yes |
| 2095 |  CHEMBL2163952 CHEMBL2163952 | C30H32N2O3 | 468.597 | 4 / 1 | 5.9 | No |
| 2358 | 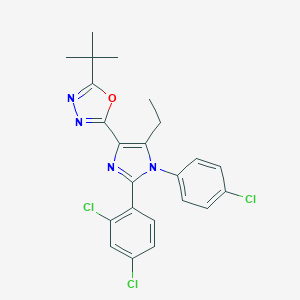 CHEMBL519280 CHEMBL519280 | C23H21Cl3N4O | 475.798 | 4 / 0 | 7.3 | No |
| 3012 | 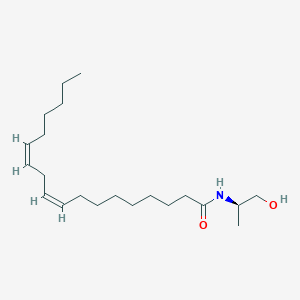 CHEMBL356213 CHEMBL356213 | C21H39NO2 | 337.548 | 2 / 2 | 6.1 | No |
| 3014 | 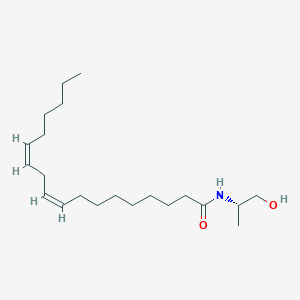 CHEMBL352931 CHEMBL352931 | C21H39NO2 | 337.548 | 2 / 2 | 6.1 | No |
| 3045 | 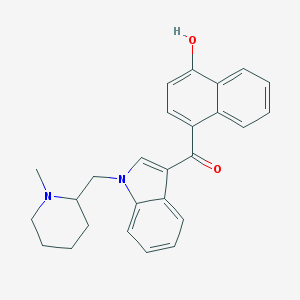 CHEMBL371328 CHEMBL371328 | C26H26N2O2 | 398.506 | 3 / 1 | 5.1 | No |
| 3073 |  CHEMBL2163572 CHEMBL2163572 | C29H38N4O3 | 490.648 | 6 / 1 | 4.2 | Yes |
| 3158 |  AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3476 |  AC1LEVMS AC1LEVMS | C17H13NO2 | 263.296 | 3 / 0 | 3.7 | Yes |
| 3519 |  CHEMBL263632 CHEMBL263632 | C19H30O2 | 290.447 | 2 / 2 | 3.8 | Yes |
| 3647 |  CHEMBL598095 CHEMBL598095 | C25H23Cl3N6O2 | 545.849 | 5 / 2 | 5.4 | No |
| 4053 |  CHEMBL149691 CHEMBL149691 | C24H43NO2 | 377.613 | 3 / 3 | 6.1 | No |
| 4065 |  AC1LP5MH AC1LP5MH | C19H16N2O2S | 336.409 | 4 / 0 | 4.1 | Yes |
| 4195 |  CHEMBL2316259 CHEMBL2316259 | C25H34N2O2S | 426.619 | 4 / 1 | 6.4 | No |
| 4260 | 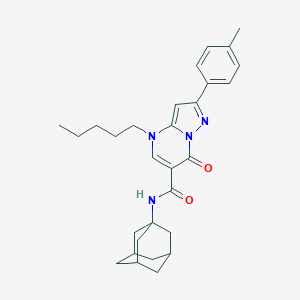 CHEMBL2381791 CHEMBL2381791 | C29H36N4O2 | 472.633 | 4 / 1 | 6.4 | No |
| 4458 | 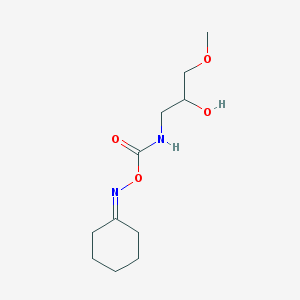 CHEMBL2172461 CHEMBL2172461 | C11H20N2O4 | 244.291 | 5 / 2 | 0.4 | Yes |
| 4463 | 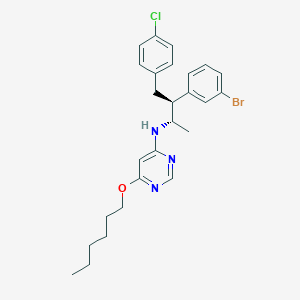 CHEMBL558725 CHEMBL558725 | C26H31BrClN3O | 516.908 | 4 / 1 | 8.6 | No |
| 5129 | 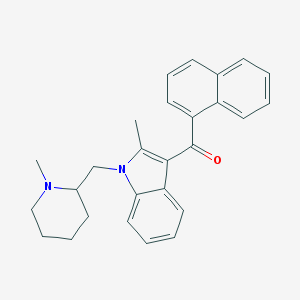 CHEMBL68642 CHEMBL68642 | C27H28N2O | 396.534 | 2 / 0 | 5.9 | No |
| 5191 |  CHEMBL557736 CHEMBL557736 | C26H24BrClN4 | 507.86 | 4 / 2 | 7.9 | No |
| 5339 |  CHEMBL2010905 CHEMBL2010905 | C19H28N2O3S | 364.504 | 4 / 2 | 3.7 | Yes |
| 6035 |  CHEMBL2316265 CHEMBL2316265 | C21H26N4O3 | 382.464 | 5 / 1 | 3.5 | Yes |
| 6476 |  CHEMBL204387 CHEMBL204387 | C24H28N2O5 | 424.497 | 5 / 3 | 4.7 | Yes |
| 6855 |  AM 281 AM 281 | C21H19Cl2IN4O2 | 557.213 | 4 / 1 | 5.3 | No |
| 7521 |  JWH-182 JWH-182 | C27H29NO | 383.535 | 1 / 0 | 7.7 | No |
| 8254 |  CHEMBL15779 CHEMBL15779 | C19H16Cl3N3O2 | 424.706 | 3 / 2 | 4.7 | Yes |
| 8535 |  SCHEMBL601645 SCHEMBL601645 | C22H27N3O | 349.478 | 2 / 0 | 4.6 | Yes |
| 8546 | 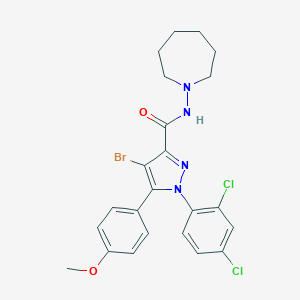 CHEMBL161163 CHEMBL161163 | C23H23BrCl2N4O2 | 538.267 | 4 / 1 | 6.5 | No |
| 8748 | 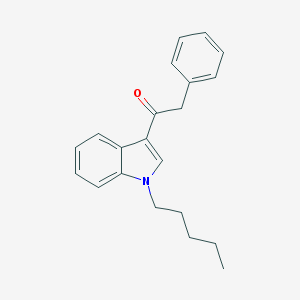 1-(1-Pentyl-1H-indol-3-yl)-2-phenylethanone 1-(1-Pentyl-1H-indol-3-yl)-2-phenylethanone | C21H23NO | 305.421 | 1 / 0 | 5.0 | Yes |
| 8851 | 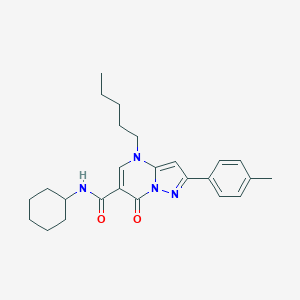 CHEMBL2381789 CHEMBL2381789 | C25H32N4O2 | 420.557 | 4 / 1 | 5.6 | No |
| 9099 | 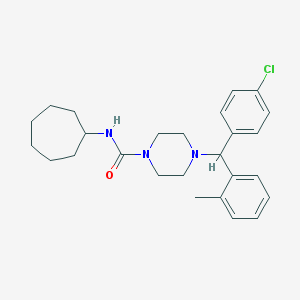 CHEMBL488031 CHEMBL488031 | C26H34ClN3O | 440.028 | 2 / 1 | 5.9 | No |
| 9158 |  CHEMBL158367 CHEMBL158367 | C25H43NO2 | 389.624 | 2 / 1 | 6.4 | No |
| 9630 |  CHEMBL2237740 CHEMBL2237740 | C17H12Cl3N3O2 | 396.652 | 4 / 0 | 5.5 | No |
| 9841 |  CHEMBL2448132 CHEMBL2448132 | C26H26N2O2 | 398.506 | 3 / 0 | 4.6 | Yes |
| 9842 |  CHEMBL2111962 CHEMBL2111962 | C26H26N2O2 | 398.506 | 3 / 0 | 4.6 | Yes |
| 9843 |  CHEMBL319464 CHEMBL319464 | C26H26N2O2 | 398.506 | 3 / 0 | 4.6 | Yes |
| 9920 |  BDBM85802 BDBM85802 | C18H17NO | 263.34 | 1 / 0 | 4.1 | Yes |
| 10022 |  CHEMBL322112 CHEMBL322112 | C29H36O2S2 | 480.725 | 4 / 1 | 8.4 | No |
| 10472 |  CHEMBL470199 CHEMBL470199 | C18H16O3 | 280.323 | 3 / 0 | 4.2 | Yes |
| 10501 | 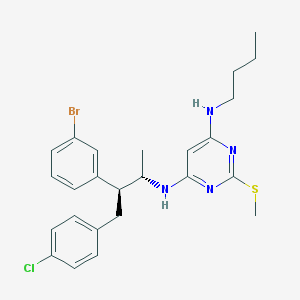 CHEMBL560747 CHEMBL560747 | C25H30BrClN4S | 533.957 | 5 / 2 | 8.4 | No |
| 10891 | 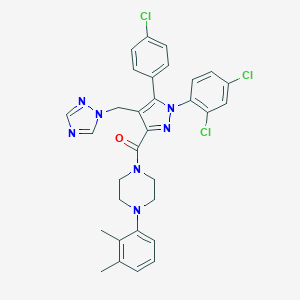 CHEMBL600682 CHEMBL600682 | C31H28Cl3N7O | 620.963 | 5 / 0 | 7.2 | No |
| 10973 | 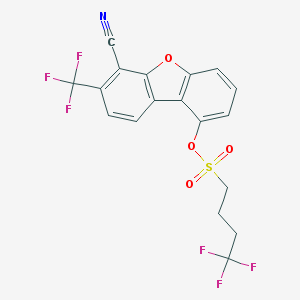 CHEMBL1834522 CHEMBL1834522 | C18H11F6NO4S | 451.339 | 11 / 0 | 5.6 | No |
| 11007 | 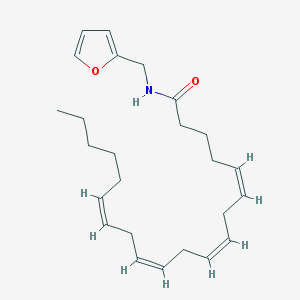 CHEMBL39460 CHEMBL39460 | C25H37NO2 | 383.576 | 2 / 1 | 6.6 | No |
| 11124 |  CHEMBL423602 CHEMBL423602 | C25H43NO | 373.625 | 1 / 1 | 7.8 | No |
| 11268 |  CHEMBL3221194 CHEMBL3221194 | C15H14N2OS | 270.35 | 2 / 0 | 3.2 | Yes |
| 442151 |  CHEMBL2113102 CHEMBL2113102 | C23H36N4O | 384.568 | 3 / 1 | 6.6 | No |
| 11660 |  CHEMBL2163571 CHEMBL2163571 | C28H35N3O3 | 461.606 | 5 / 1 | 5.2 | No |
| 12000 | 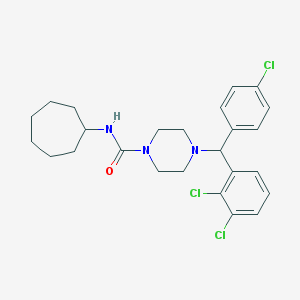 CHEMBL472300 CHEMBL472300 | C25H30Cl3N3O | 494.885 | 2 / 1 | 6.8 | No |
| 12205 | 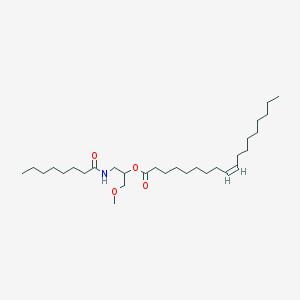 CHEMBL2172457 CHEMBL2172457 | C30H57NO4 | 495.789 | 4 / 1 | 9.8 | No |
| 12258 | 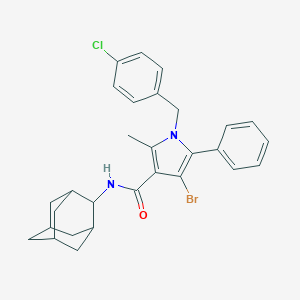 CHEMBL519302 CHEMBL519302 | C29H30BrClN2O | 537.926 | 1 / 1 | 7.3 | No |
| 12511 | 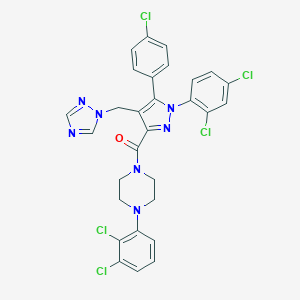 CHEMBL600480 CHEMBL600480 | C29H22Cl5N7O | 661.793 | 5 / 0 | 7.8 | No |
| 12781 |  CHEMBL419138 CHEMBL419138 | C20H19Cl3N4O | 437.749 | 3 / 2 | 6.1 | No |
| 12801 |  CHEMBL2316278 CHEMBL2316278 | C26H33ClN4O2 | 469.026 | 4 / 1 | 6.3 | No |
| 12922 |  CHEMBL1223179 CHEMBL1223179 | C25H18Cl2F3N7S | 576.423 | 9 / 0 | 6.9 | No |
| 13714 |  CHEMBL606380 CHEMBL606380 | C29H24BrCl2N7O | 637.363 | 5 / 0 | 6.6 | No |
| 13922 | 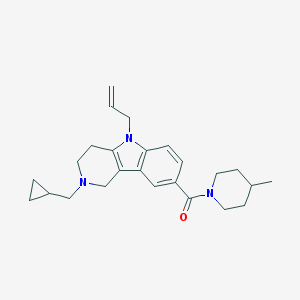 CHEMBL1950330 CHEMBL1950330 | C25H33N3O | 391.559 | 2 / 0 | 4.1 | Yes |
| 14567 | 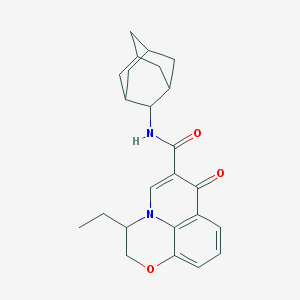 CHEMBL2163930 CHEMBL2163930 | C24H28N2O3 | 392.499 | 4 / 1 | 4.3 | Yes |
| 16124 | 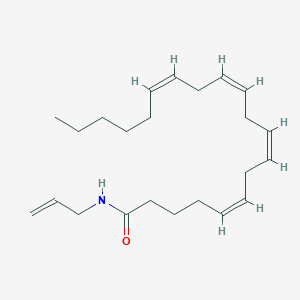 CHEMBL274016 CHEMBL274016 | C23H37NO | 343.555 | 1 / 1 | 6.7 | No |
| 16271 | 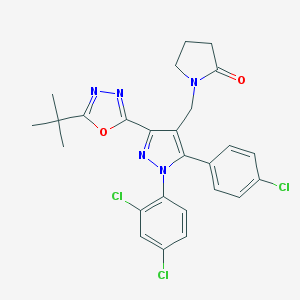 CHEMBL487583 CHEMBL487583 | C26H24Cl3N5O2 | 544.861 | 5 / 0 | 6.0 | No |
| 16417 |  CHEMBL497557 CHEMBL497557 | C25H25ClF3N5O | 503.954 | 8 / 1 | 4.9 | No |
| 16749 |  CHEMBL346994 CHEMBL346994 | C23H20Cl2N4O2S | 487.399 | 4 / 1 | 5.3 | No |
| 442322 |  Ibipinabant Ibipinabant | C23H20Cl2N4O2S | 487.399 | 4 / 1 | 5.3 | No |
| 442323 |  362519-49-1 362519-49-1 | C23H20Cl2N4O2S | 487.399 | 4 / 1 | 5.3 | No |
| 16834 | 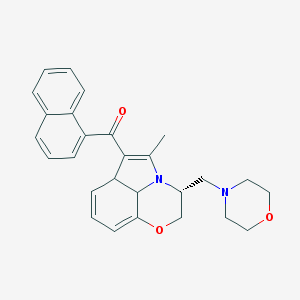 BDBM82141 BDBM82141 | C27H28N2O3 | 428.532 | 5 / 0 | 3.8 | Yes |
| 16890 | 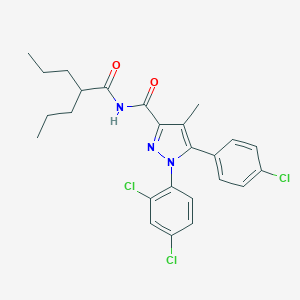 CHEMBL520472 CHEMBL520472 | C25H26Cl3N3O2 | 506.852 | 3 / 1 | 8.0 | No |
| 17181 | 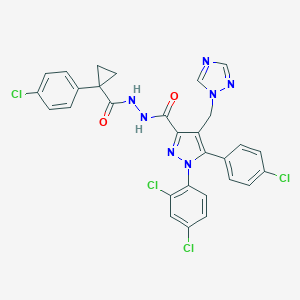 CHEMBL589446 CHEMBL589446 | C29H21Cl4N7O2 | 641.334 | 5 / 2 | 6.8 | No |
| 17745 | 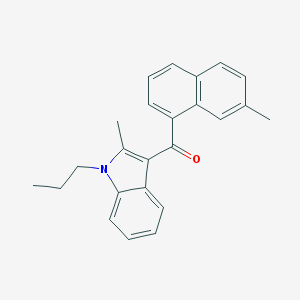 CHEMBL558801 CHEMBL558801 | C24H23NO | 341.454 | 1 / 0 | 6.2 | No |
| 17830 |  CHEMBL1223177 CHEMBL1223177 | C24H21Cl2N7S | 510.441 | 6 / 0 | 6.3 | No |
| 18282 |  CHEMBL500927 CHEMBL500927 | C19H12ClF4N5O3 | 469.781 | 11 / 1 | 3.7 | No |
| 18307 |  CHEMBL483944 CHEMBL483944 | C21H18Cl3N3O2 | 450.744 | 3 / 1 | 6.2 | No |
| 18365 |  CHEMBL551869 CHEMBL551869 | C16H9Cl2NO | 302.154 | 2 / 0 | 5.1 | No |
| 18412 |  CHEMBL487173 CHEMBL487173 | C25H22Cl3N5O3 | 546.833 | 6 / 0 | 6.1 | No |
| 18535 |  CHEMBL563081 CHEMBL563081 | C25H26Cl2N4O | 469.41 | 3 / 1 | 6.8 | No |
| 19271 |  CHEMBL1269764 CHEMBL1269764 | C38H31Cl6N7O2 | 830.413 | 5 / 3 | 10.2 | No |
| 19457 |  CHEMBL2316281 CHEMBL2316281 | C23H31N3O3 | 397.519 | 5 / 1 | 4.8 | Yes |
| 19522 | 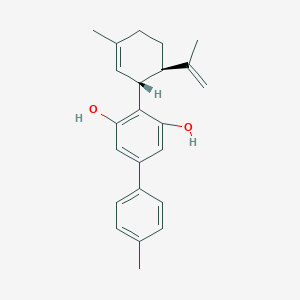 CHEMBL205685 CHEMBL205685 | C23H26O2 | 334.459 | 2 / 2 | 6.1 | No |
| 20321 | 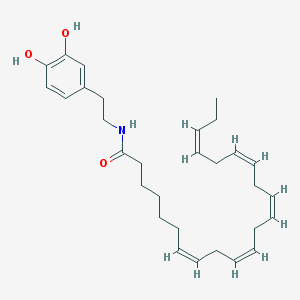 CHEMBL422633 CHEMBL422633 | C30H43NO3 | 465.678 | 3 / 3 | 7.7 | No |
| 20449 | 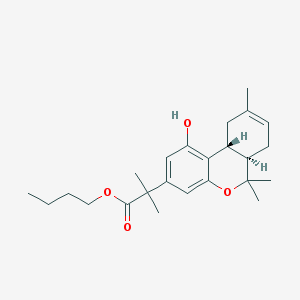 CHEMBL3104360 CHEMBL3104360 | C24H34O4 | 386.532 | 4 / 1 | 5.3 | No |
| 20741 | 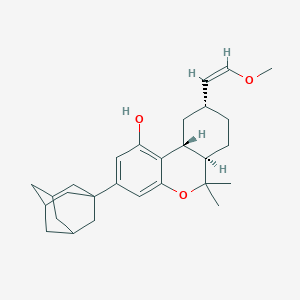 CHEMBL2348465 CHEMBL2348465 | C28H38O3 | 422.609 | 3 / 1 | 7.4 | No |
| 21209 | 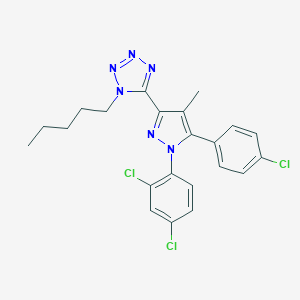 CHEMBL272507 CHEMBL272507 | C22H21Cl3N6 | 475.802 | 4 / 0 | 6.9 | No |
| 21412 | 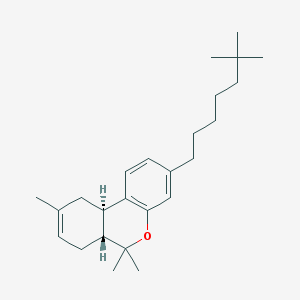 CHEMBL2112646 CHEMBL2112646 | C25H38O | 354.578 | 1 / 0 | 7.8 | No |
| 21638 | 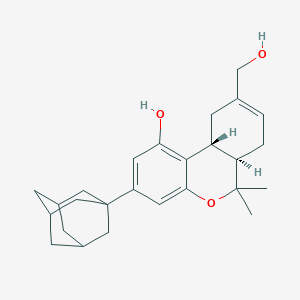 CHEMBL2348472 CHEMBL2348472 | C26H34O3 | 394.555 | 3 / 2 | 5.5 | No |
| 21682 | 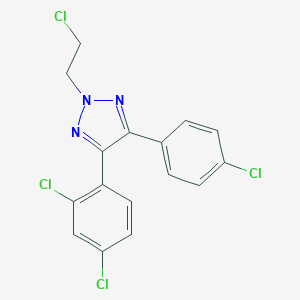 CHEMBL2237748 CHEMBL2237748 | C16H11Cl4N3 | 387.085 | 2 / 0 | 6.0 | No |
| 21965 | 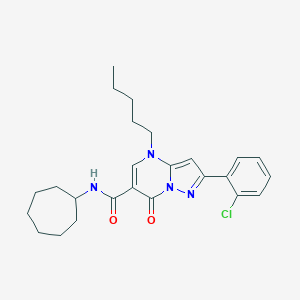 CHEMBL2381793 CHEMBL2381793 | C25H31ClN4O2 | 454.999 | 4 / 1 | 6.4 | No |
| 22332 | 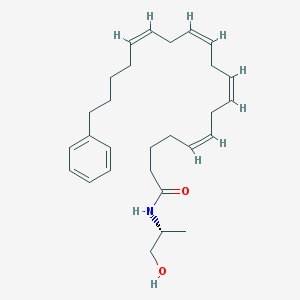 CHEMBL460307 CHEMBL460307 | C28H41NO2 | 423.641 | 2 / 2 | 6.7 | No |
| 22418 | 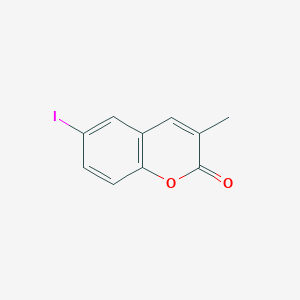 CHEMBL455557 CHEMBL455557 | C10H7IO2 | 286.068 | 2 / 0 | 2.9 | Yes |
| 22425 | 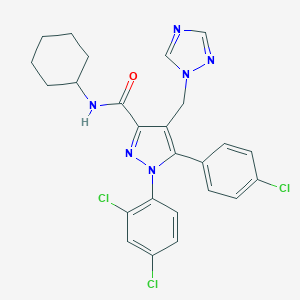 CHEMBL605757 CHEMBL605757 | C25H23Cl3N6O | 529.85 | 4 / 1 | 6.5 | No |
| 23403 | 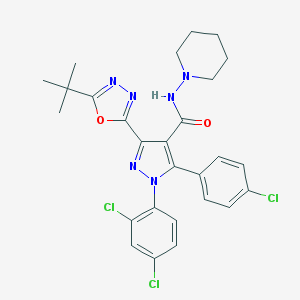 CHEMBL485153 CHEMBL485153 | C27H27Cl3N6O2 | 573.903 | 6 / 1 | 7.0 | No |
| 23450 | 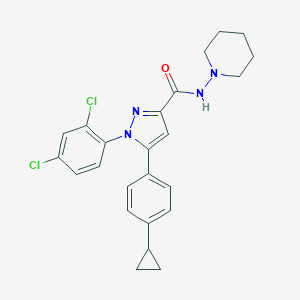 CHEMBL556686 CHEMBL556686 | C24H24Cl2N4O | 455.383 | 3 / 1 | 6.4 | No |
| 24216 | 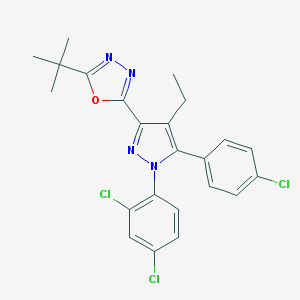 CHEMBL486756 CHEMBL486756 | C23H21Cl3N4O | 475.798 | 4 / 0 | 7.5 | No |
| 24298 | 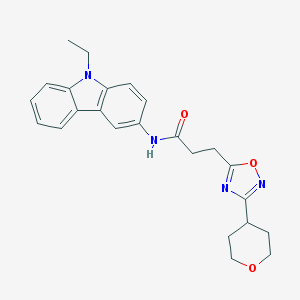 CHEMBL500655 CHEMBL500655 | C24H26N4O3 | 418.497 | 5 / 1 | 3.4 | Yes |
| 24357 | 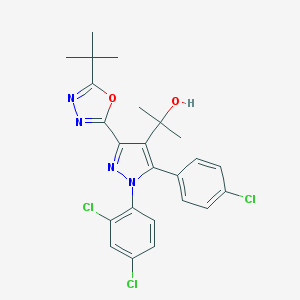 CHEMBL519875 CHEMBL519875 | C24H23Cl3N4O2 | 505.824 | 5 / 1 | 6.4 | No |
| 24423 | 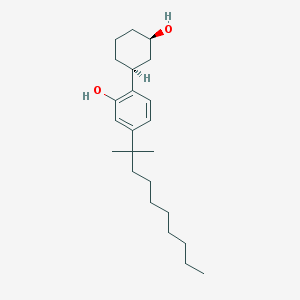 UNII-7W96Y4OF1O UNII-7W96Y4OF1O | C23H38O2 | 346.555 | 2 / 2 | 8.2 | No |
| 442628 | 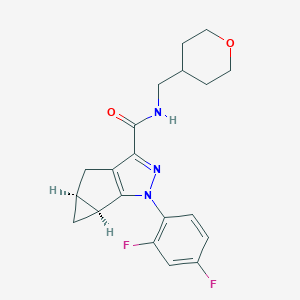 CHEMBL3354966 CHEMBL3354966 | C20H21F2N3O2 | 373.404 | 5 / 1 | 2.8 | Yes |
| 442634 | 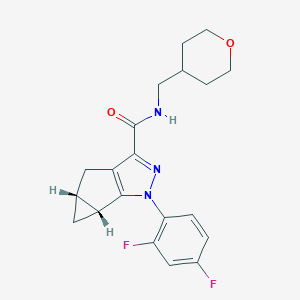 CHEMBL3354959 CHEMBL3354959 | C20H21F2N3O2 | 373.404 | 5 / 1 | 2.8 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417