

You can:
| Name | N-formyl peptide receptor 3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | FPR3 |
| Synonym | FMLP-related receptor II Lxa4r LXA4-R FPRL2 Fprl1 [ Show all ] |
| Disease | N/A |
| Length | 353 |
| Amino acid sequence | METNFSIPLNETEEVLPEPAGHTVLWIFSLLVHGVTFVFGVLGNGLVIWVAGFRMTRTVNTICYLNLALADFSFSAILPFRMVSVAMREKWPFGSFLCKLVHVMIDINLFVSVYLITIIALDRCICVLHPAWAQNHRTMSLAKRVMTGLWIFTIVLTLPNFIFWTTISTTNGDTYCIFNFAFWGDTAVERLNVFITMAKVFLILHFIIGFSVPMSIITVCYGIIAAKIHRNHMIKSSRPLRVFAAVVASFFICWFPYELIGILMAVWLKEMLLNGKYKIILVLINPTSSLAFFNSCLNPILYVFMGRNFQERLIRSLPTSLERALTEVPDSAQTSNTDTTSASPPEETELQAM |
| UniProt | P25089 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P25089 |
| 3D structure model | This predicted structure model is from GPCR-EXP P25089. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | N/A |
| IUPHAR | 224 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 498 | 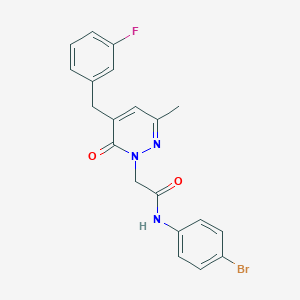 CHEMBL2391280 CHEMBL2391280 | C20H17BrFN3O2 | 430.277 | 4 / 1 | 3.4 | Yes |
| 7082 | 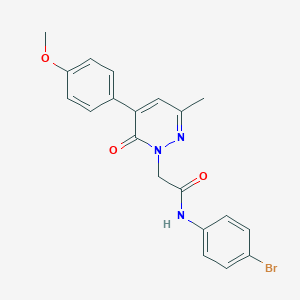 CHEMBL2391452 CHEMBL2391452 | C20H18BrN3O3 | 428.286 | 4 / 1 | 3.0 | Yes |
| 13260 | 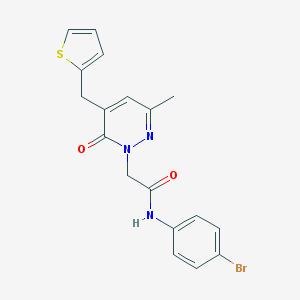 CHEMBL2391437 CHEMBL2391437 | C18H16BrN3O2S | 418.309 | 4 / 1 | 3.0 | Yes |
| 38566 | 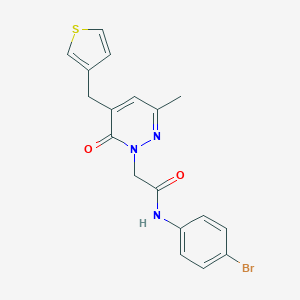 CHEMBL2391286 CHEMBL2391286 | C18H16BrN3O2S | 418.309 | 4 / 1 | 3.0 | Yes |
| 46714 | 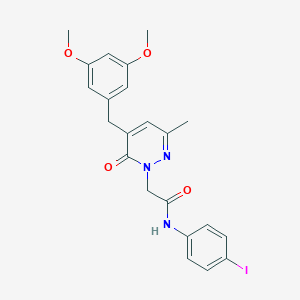 CHEMBL2391442 CHEMBL2391442 | C22H22IN3O4 | 519.339 | 5 / 1 | 3.2 | No |
| 49232 | 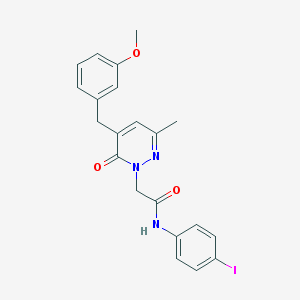 CHEMBL560780 CHEMBL560780 | C21H20IN3O3 | 489.313 | 4 / 1 | 3.2 | Yes |
| 55865 | 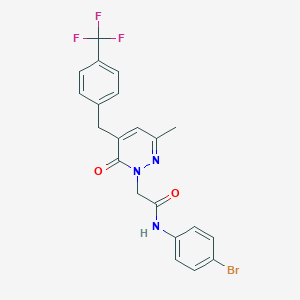 CHEMBL2391284 CHEMBL2391284 | C21H17BrF3N3O2 | 480.285 | 6 / 1 | 4.2 | Yes |
| 58538 | 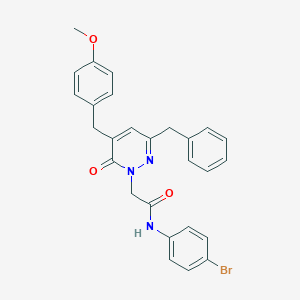 CHEMBL2391276 CHEMBL2391276 | C27H24BrN3O3 | 518.411 | 4 / 1 | 4.9 | No |
| 60268 | 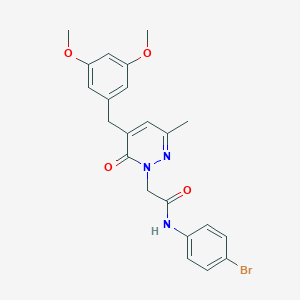 CHEMBL2391283 CHEMBL2391283 | C22H22BrN3O4 | 472.339 | 5 / 1 | 3.3 | Yes |
| 553578 | 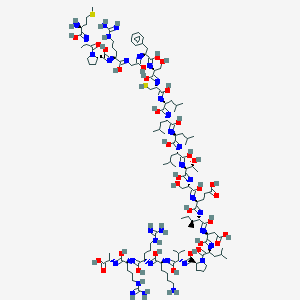 CID 56947109 CID 56947109 | C119H204N34O32S2 | 2687.27 | 60 / 39 | 4.5 | No |
| 81775 | 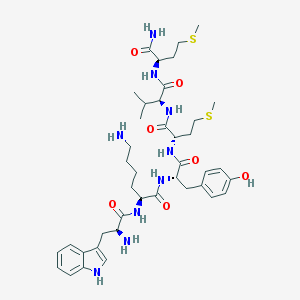 WKYMVm WKYMVm | C41H61N9O7S2 | 856.115 | 11 / 10 | 1.5 | No |
| 83585 | 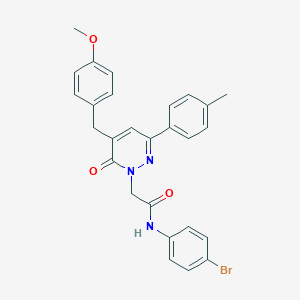 CHEMBL2391270 CHEMBL2391270 | C27H24BrN3O3 | 518.411 | 4 / 1 | 5.3 | No |
| 84651 | 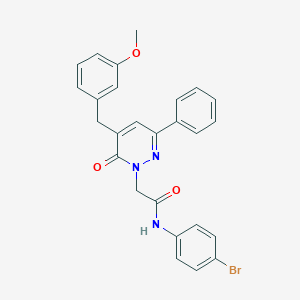 CHEMBL2391258 CHEMBL2391258 | C26H22BrN3O3 | 504.384 | 4 / 1 | 4.9 | No |
| 102524 | 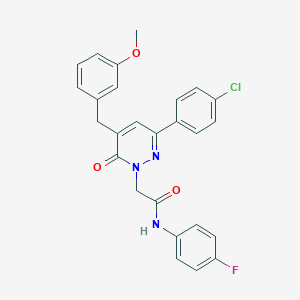 CHEMBL2391269 CHEMBL2391269 | C26H21ClFN3O3 | 477.92 | 5 / 1 | 5.0 | Yes |
| 119938 | 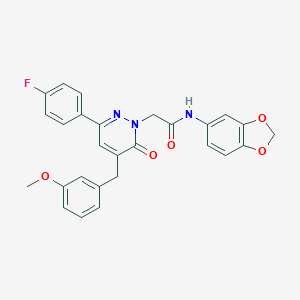 CHEMBL2391274 CHEMBL2391274 | C27H22FN3O5 | 487.487 | 7 / 1 | 4.2 | Yes |
| 126891 | 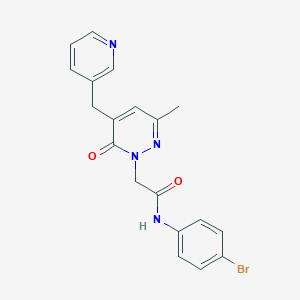 CHEMBL2391439 CHEMBL2391439 | C19H17BrN4O2 | 413.275 | 4 / 1 | 2.2 | Yes |
| 134547 | 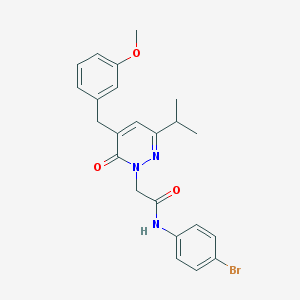 CHEMBL2391256 CHEMBL2391256 | C23H24BrN3O3 | 470.367 | 4 / 1 | 4.3 | Yes |
| 136887 | 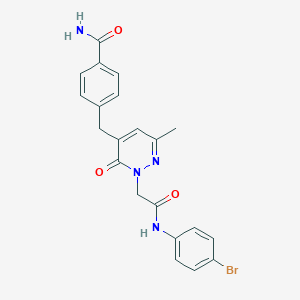 CHEMBL2391440 CHEMBL2391440 | C21H19BrN4O3 | 455.312 | 4 / 2 | 2.2 | Yes |
| 138444 | 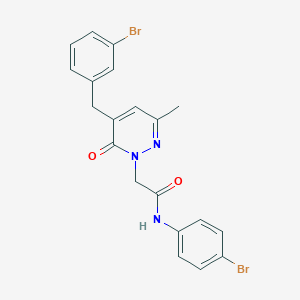 CHEMBL2391282 CHEMBL2391282 | C20H17Br2N3O2 | 491.183 | 3 / 1 | 4.0 | Yes |
| 163630 | 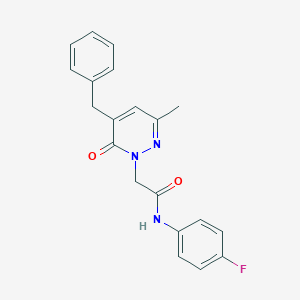 CHEMBL2391278 CHEMBL2391278 | C20H18FN3O2 | 351.381 | 4 / 1 | 2.7 | Yes |
| 175999 | 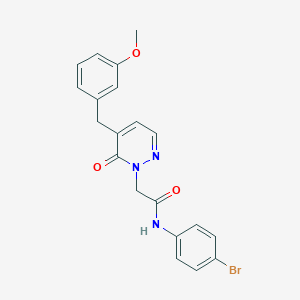 CHEMBL2391254 CHEMBL2391254 | C20H18BrN3O3 | 428.286 | 4 / 1 | 3.0 | Yes |
| 186218 | 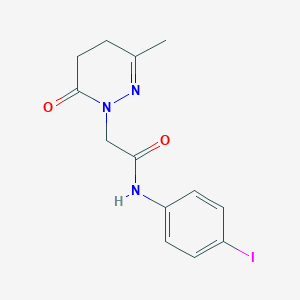 CHEMBL2391444 CHEMBL2391444 | C13H14IN3O2 | 371.178 | 3 / 1 | 1.0 | Yes |
| 188576 | 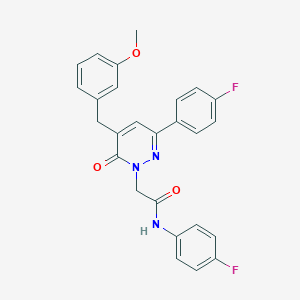 CHEMBL2391275 CHEMBL2391275 | C26H21F2N3O3 | 461.469 | 6 / 1 | 4.4 | Yes |
| 197780 | 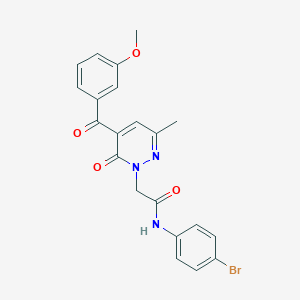 CHEMBL2391446 CHEMBL2391446 | C21H18BrN3O4 | 456.296 | 5 / 1 | 2.9 | Yes |
| 221310 | 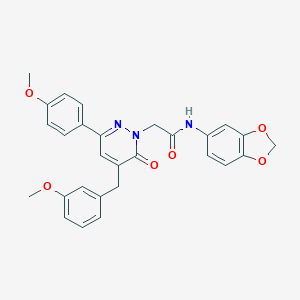 CHEMBL2391265 CHEMBL2391265 | C28H25N3O6 | 499.523 | 7 / 1 | 4.0 | Yes |
| 224217 | 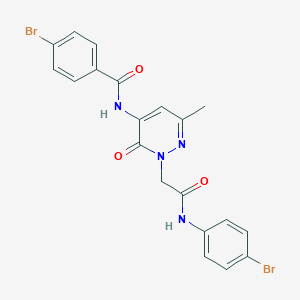 CHEMBL2391451 CHEMBL2391451 | C20H16Br2N4O3 | 520.181 | 4 / 2 | 3.1 | No |
| 234931 | 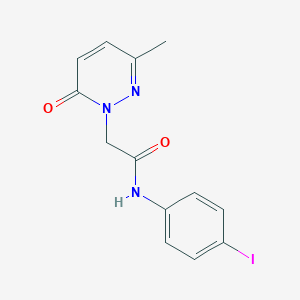 CHEMBL2391445 CHEMBL2391445 | C13H12IN3O2 | 369.162 | 3 / 1 | 1.3 | Yes |
| 244942 | 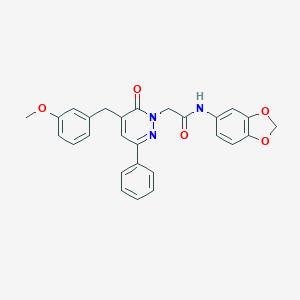 CHEMBL2391259 CHEMBL2391259 | C27H23N3O5 | 469.497 | 6 / 1 | 4.1 | Yes |
| 245568 | 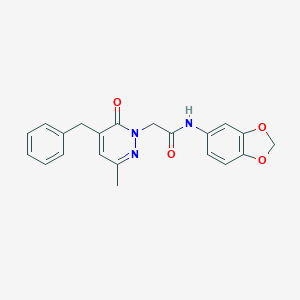 CHEMBL2391279 CHEMBL2391279 | C21H19N3O4 | 377.4 | 5 / 1 | 2.4 | Yes |
| 260441 | 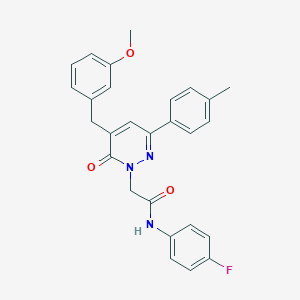 CHEMBL2391272 CHEMBL2391272 | C27H24FN3O3 | 457.505 | 5 / 1 | 4.7 | Yes |
| 267110 | 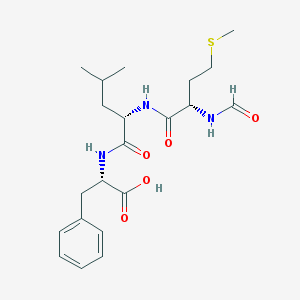 59880-97-6 59880-97-6 | C21H31N3O5S | 437.555 | 6 / 4 | 1.5 | Yes |
| 267695 | 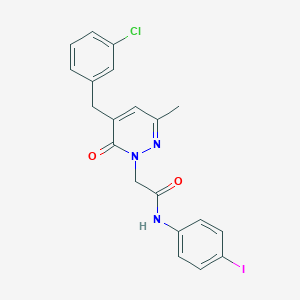 CHEMBL2391443 CHEMBL2391443 | C20H17ClIN3O2 | 493.729 | 3 / 1 | 3.9 | Yes |
| 270253 | 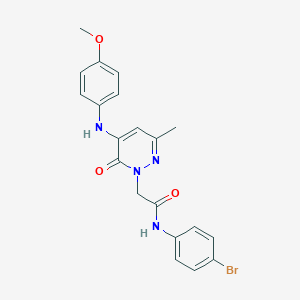 CHEMBL2391449 CHEMBL2391449 | C20H19BrN4O3 | 443.301 | 5 / 2 | 3.1 | Yes |
| 276783 | 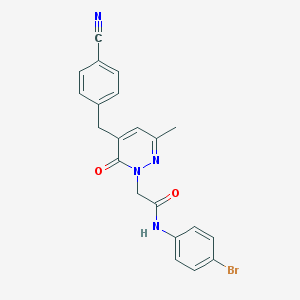 CHEMBL2391441 CHEMBL2391441 | C21H17BrN4O2 | 437.297 | 4 / 1 | 3.0 | Yes |
| 282289 | 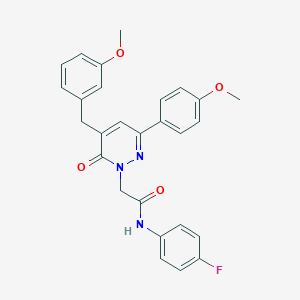 CHEMBL2391266 CHEMBL2391266 | C27H24FN3O4 | 473.504 | 6 / 1 | 4.3 | Yes |
| 282579 | 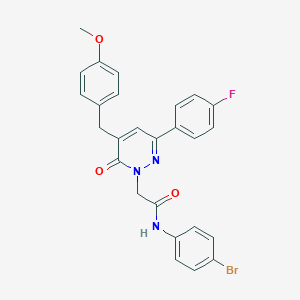 CHEMBL2391273 CHEMBL2391273 | C26H21BrFN3O3 | 522.374 | 5 / 1 | 5.0 | No |
| 283569 | 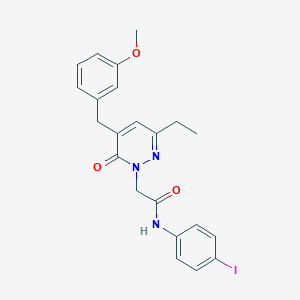 CHEMBL2391255 CHEMBL2391255 | C22H22IN3O3 | 503.34 | 4 / 1 | 3.7 | No |
| 306037 | 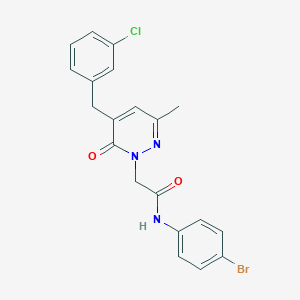 CHEMBL2391281 CHEMBL2391281 | C20H17BrClN3O2 | 446.729 | 3 / 1 | 3.9 | Yes |
| 320799 | 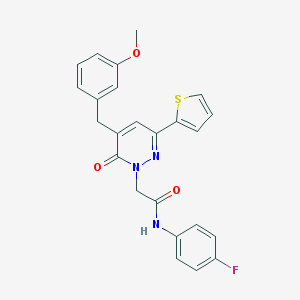 CHEMBL2391263 CHEMBL2391263 | C24H20FN3O3S | 449.5 | 6 / 1 | 4.4 | Yes |
| 326631 | 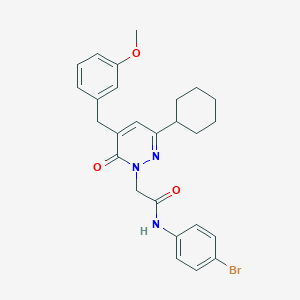 CHEMBL2391257 CHEMBL2391257 | C26H28BrN3O3 | 510.432 | 4 / 1 | 5.4 | No |
| 346536 | 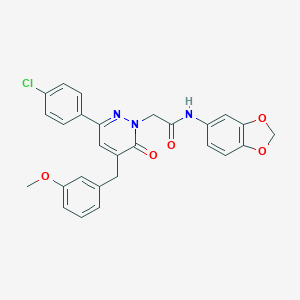 CHEMBL2391268 CHEMBL2391268 | C27H22ClN3O5 | 503.939 | 6 / 1 | 4.7 | No |
| 362151 | 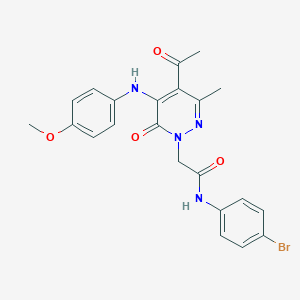 CHEMBL2391447 CHEMBL2391447 | C22H21BrN4O4 | 485.338 | 6 / 2 | 3.5 | Yes |
| 362502 | 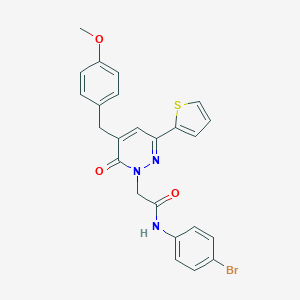 CHEMBL2391261 CHEMBL2391261 | C24H20BrN3O3S | 510.406 | 5 / 1 | 5.0 | No |
| 370994 | 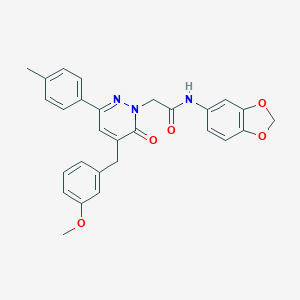 CHEMBL2391271 CHEMBL2391271 | C28H25N3O5 | 483.524 | 6 / 1 | 4.4 | Yes |
| 379498 | 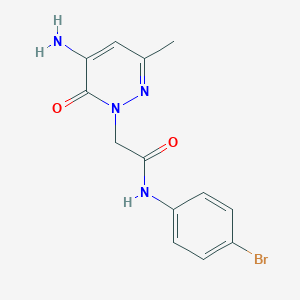 CHEMBL2391448 CHEMBL2391448 | C13H13BrN4O2 | 337.177 | 4 / 2 | 0.9 | Yes |
| 382695 | 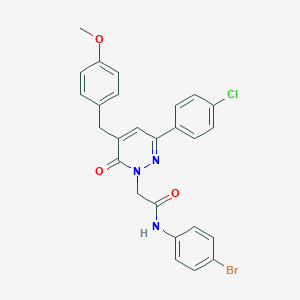 CHEMBL2391267 CHEMBL2391267 | C26H21BrClN3O3 | 538.826 | 4 / 1 | 5.6 | No |
| 403863 | 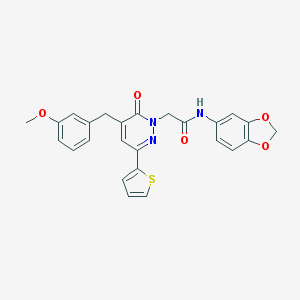 CHEMBL2391262 CHEMBL2391262 | C25H21N3O5S | 475.519 | 7 / 1 | 4.1 | Yes |
| 412526 | 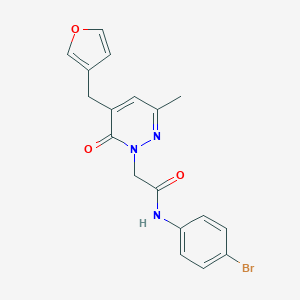 CHEMBL2391285 CHEMBL2391285 | C18H16BrN3O3 | 402.248 | 4 / 1 | 2.4 | Yes |
| 418547 | 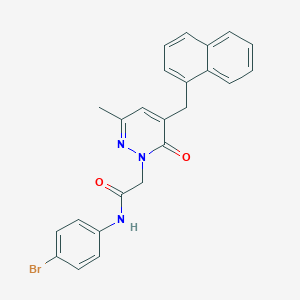 CHEMBL2391438 CHEMBL2391438 | C24H20BrN3O2 | 462.347 | 3 / 1 | 4.6 | Yes |
| 428756 | 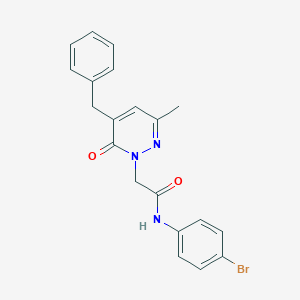 CHEMBL2391277 CHEMBL2391277 | C20H18BrN3O2 | 412.287 | 3 / 1 | 3.3 | Yes |
| 431014 | 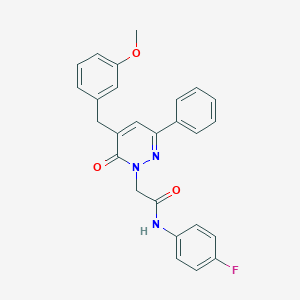 CHEMBL2391260 CHEMBL2391260 | C26H22FN3O3 | 443.478 | 5 / 1 | 4.3 | Yes |
| 439065 | 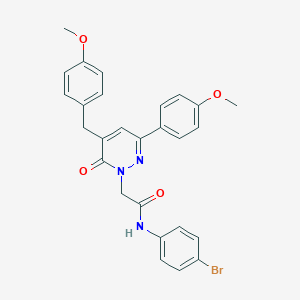 CHEMBL2391264 CHEMBL2391264 | C27H24BrN3O4 | 534.41 | 5 / 1 | 4.9 | No |
| 440292 | 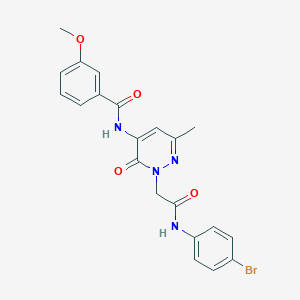 CHEMBL2391450 CHEMBL2391450 | C21H19BrN4O4 | 471.311 | 5 / 2 | 2.4 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417