

You can:
| Name | C3a anaphylatoxin chemotactic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | C3AR1 |
| Synonym | C3a anaphylatoxin chemotactic receptor C3a receptor C3AR anaphylatoxin C3a receptor complement component 3a receptor 1 [ Show all ] |
| Disease | N/A |
| Length | 482 |
| Amino acid sequence | MASFSAETNSTDLLSQPWNEPPVILSMVILSLTFLLGLPGNGLVLWVAGLKMQRTVNTIWFLHLTLADLLCCLSLPFSLAHLALQGQWPYGRFLCKLIPSIIVLNMFASVFLLTAISLDRCLVVFKPIWCQNHRNVGMACSICGCIWVVAFVMCIPVFVYREIFTTDNHNRCGYKFGLSSSLDYPDFYGDPLENRSLENIVQPPGEMNDRLDPSSFQTNDHPWTVPTVFQPQTFQRPSADSLPRGSARLTSQNLYSNVFKPADVVSPKIPSGFPIEDHETSPLDNSDAFLSTHLKLFPSASSNSFYESELPQGFQDYYNLGQFTDDDQVPTPLVAITITRLVVGFLLPSVIMIACYSFIVFRMQRGRFAKSQSKTFRVAVVVVAVFLVCWTPYHIFGVLSLLTDPETPLGKTLMSWDHVCIALASANSCFNPFLYALLGKDFRKKARQSIQGILEAAFSEELTRSTHCPSNNVISERNSTTV |
| UniProt | Q16581 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q16581 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q16581. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4761 |
| IUPHAR | 31 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 651 | 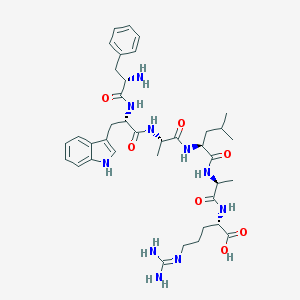 CHEMBL1170635 CHEMBL1170635 | C38H54N10O7 | 762.913 | 9 / 10 | -1.6 | No |
| 521471 | 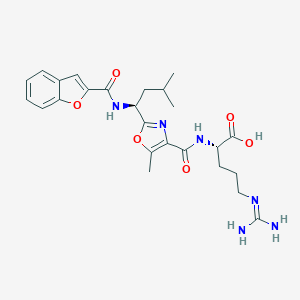 CHEMBL3736483 CHEMBL3736483 | C25H32N6O6 | 512.567 | 8 / 5 | 2.4 | No |
| 4721 | 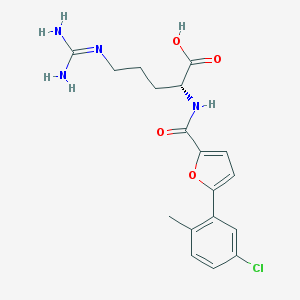 CHEMBL228582 CHEMBL228582 | C18H21ClN4O4 | 392.84 | 5 / 4 | 2.0 | Yes |
| 521613 | 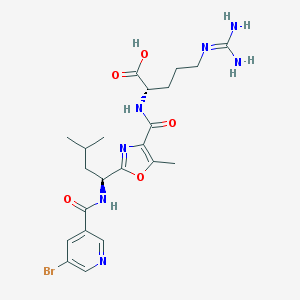 CHEMBL3736513 CHEMBL3736513 | C22H30BrN7O5 | 552.43 | 8 / 5 | 1.3 | No |
| 6391 | 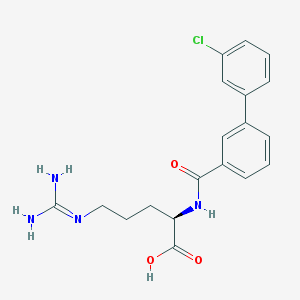 CHEMBL228847 CHEMBL228847 | C19H21ClN4O3 | 388.852 | 4 / 4 | 2.3 | Yes |
| 15073 | 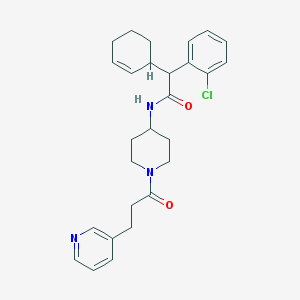 CHEMBL234100 CHEMBL234100 | C27H32ClN3O2 | 466.022 | 3 / 1 | 4.7 | Yes |
| 442412 | 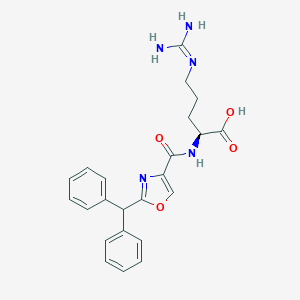 CHEMBL3342685 CHEMBL3342685 | C23H25N5O4 | 435.484 | 6 / 4 | 2.2 | Yes |
| 19928 | 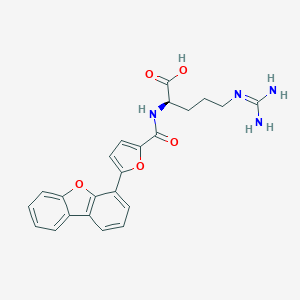 CHEMBL228635 CHEMBL228635 | C23H22N4O5 | 434.452 | 6 / 4 | 2.8 | Yes |
| 24022 | 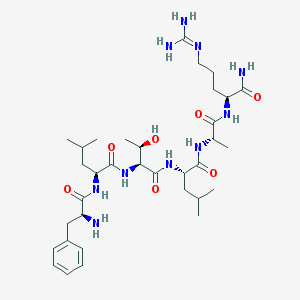 CHEMBL1170650 CHEMBL1170650 | C34H58N10O7 | 718.901 | 9 / 10 | -0.2 | No |
| 25524 | 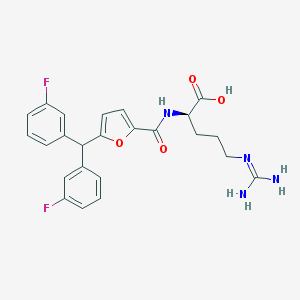 CHEMBL229005 CHEMBL229005 | C24H24F2N4O4 | 470.477 | 7 / 4 | 3.0 | Yes |
| 26804 | 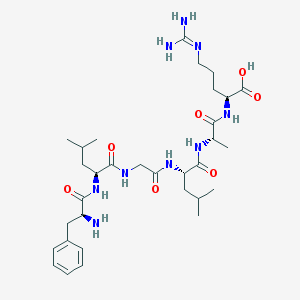 CHEMBL1170639 CHEMBL1170639 | C32H53N9O7 | 675.832 | 9 / 9 | -2.1 | No |
| 27418 | 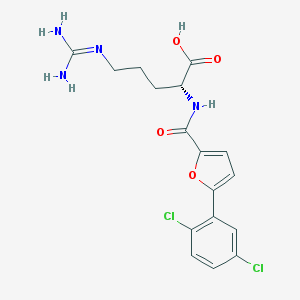 CHEMBL387686 CHEMBL387686 | C17H18Cl2N4O4 | 413.255 | 5 / 4 | 2.3 | Yes |
| 522511 | 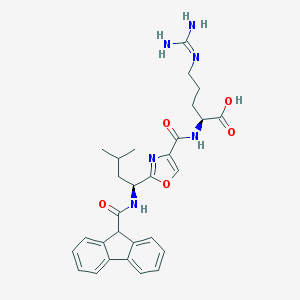 CHEMBL3734765 CHEMBL3734765 | C29H34N6O5 | 546.628 | 7 / 5 | 2.6 | No |
| 40086 | 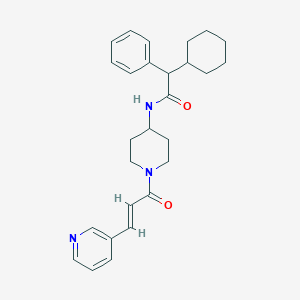 CHEMBL234523 CHEMBL234523 | C27H33N3O2 | 431.58 | 3 / 1 | 5.0 | Yes |
| 41520 | 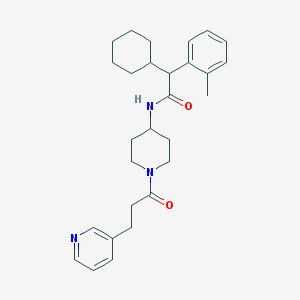 CHEMBL232029 CHEMBL232029 | C28H37N3O2 | 447.623 | 3 / 1 | 5.1 | No |
| 43841 | 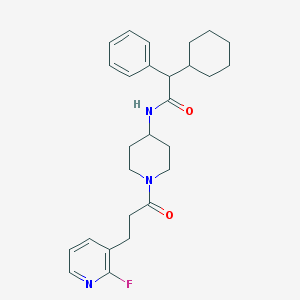 CHEMBL396031 CHEMBL396031 | C27H34FN3O2 | 451.586 | 4 / 1 | 5.2 | No |
| 48132 |  CHEMBL233232 CHEMBL233232 | C28H35N3O2 | 445.607 | 3 / 1 | 5.0 | Yes |
| 522979 |  CHEMBL3735746 CHEMBL3735746 | C24H34N6O7 | 518.571 | 9 / 5 | 1.2 | No |
| 53228 |  CHEMBL231405 CHEMBL231405 | C17H19ClN4O4 | 378.813 | 5 / 4 | 1.7 | Yes |
| 53644 |  CHEMBL1170025 CHEMBL1170025 | C76H123N17O20S2 | 1659.04 | 24 / 21 | 0.3 | No |
| 443887 | 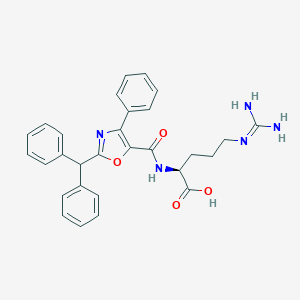 CHEMBL3342683 CHEMBL3342683 | C29H29N5O4 | 511.582 | 6 / 4 | 3.8 | No |
| 74381 | 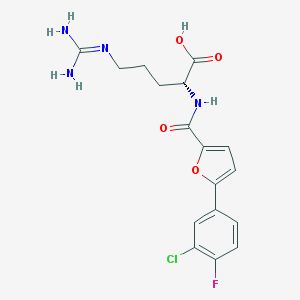 CHEMBL265866 CHEMBL265866 | C17H18ClFN4O4 | 396.803 | 6 / 4 | 1.8 | Yes |
| 523750 | 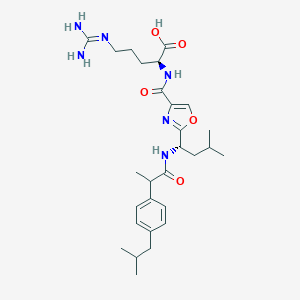 CHEMBL3736489 CHEMBL3736489 | C28H42N6O5 | 542.681 | 7 / 5 | 3.4 | No |
| 76848 | 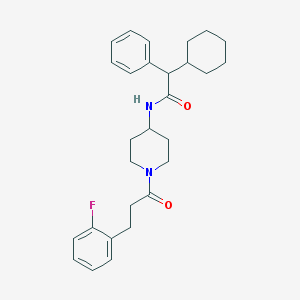 CHEMBL396262 CHEMBL396262 | C28H35FN2O2 | 450.598 | 3 / 1 | 6.0 | No |
| 80472 |  CHEMBL234731 CHEMBL234731 | C28H33FN2O2 | 448.582 | 3 / 1 | 6.1 | No |
| 86020 |  CHEMBL1170229 CHEMBL1170229 | C43H63N11O7 | 846.047 | 10 / 10 | -1.1 | No |
| 524026 |  CHEMBL3735774 CHEMBL3735774 | C23H32N6O7S | 536.604 | 9 / 5 | 1.2 | No |
| 90740 |  CHEMBL94579 CHEMBL94579 | C27H29N3O | 411.549 | 3 / 1 | 5.6 | No |
| 524412 | 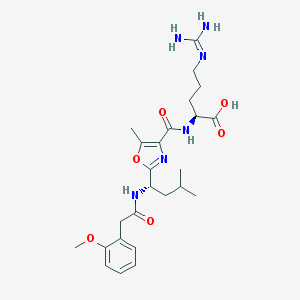 CHEMBL3734890 CHEMBL3734890 | C25H36N6O6 | 516.599 | 8 / 5 | 1.6 | No |
| 100407 | 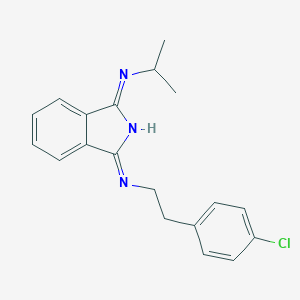 CHEMBL96477 CHEMBL96477 | C19H20ClN3 | 325.84 | 2 / 1 | 4.3 | Yes |
| 103654 | 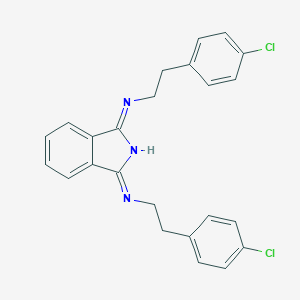 CHEMBL313866 CHEMBL313866 | C24H21Cl2N3 | 422.353 | 2 / 1 | 6.1 | No |
| 120042 | 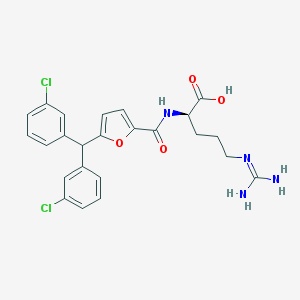 CHEMBL387755 CHEMBL387755 | C24H24Cl2N4O4 | 503.38 | 5 / 4 | 4.1 | No |
| 121212 | 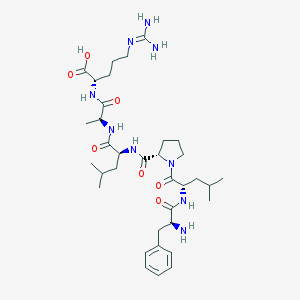 CHEMBL1172879 CHEMBL1172879 | C35H57N9O7 | 715.897 | 9 / 8 | -2.4 | No |
| 525200 | 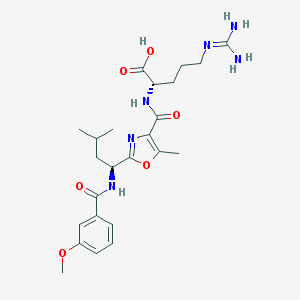 CHEMBL3736115 CHEMBL3736115 | C24H34N6O6 | 502.572 | 8 / 5 | 1.6 | No |
| 127999 | 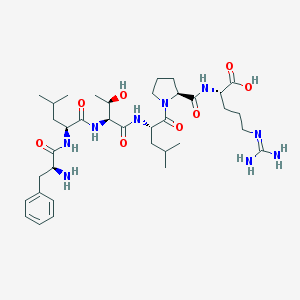 CHEMBL1170648 CHEMBL1170648 | C36H59N9O8 | 745.923 | 10 / 9 | -3.1 | No |
| 525229 | 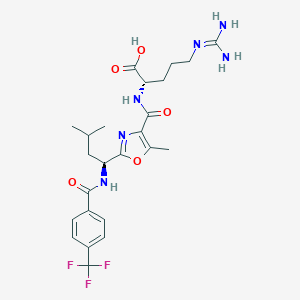 CHEMBL3736337 CHEMBL3736337 | C24H31F3N6O5 | 540.544 | 10 / 5 | 2.6 | No |
| 525256 |  CHEMBL3735240 CHEMBL3735240 | C29H38N6O6 | 566.659 | 8 / 5 | 3.0 | No |
| 129779 |  CHEMBL233028 CHEMBL233028 | C27H33F2N3O2 | 469.577 | 5 / 1 | 4.2 | Yes |
| 129783 |  CHEMBL97335 CHEMBL97335 | C24H21Cl2N3 | 422.353 | 2 / 1 | 6.1 | No |
| 130053 |  CID 70694925 CID 70694925 | C95H145N31O21 | 2057.4 | 26 / 35 | -4.2 | No |
| 525296 |  CHEMBL3736340 CHEMBL3736340 | C26H38N6O7 | 546.625 | 9 / 5 | 1.6 | No |
| 131809 |  CHEMBL234522 CHEMBL234522 | C27H34FN3O2 | 451.586 | 4 / 1 | 5.2 | No |
| 525349 |  CHEMBL3736020 CHEMBL3736020 | C26H36N6O5 | 512.611 | 7 / 5 | 2.2 | No |
| 133629 |  CHEMBL319332 CHEMBL319332 | C24H23N3 | 353.469 | 2 / 1 | 5.0 | Yes |
| 134530 |  CHEMBL1170349 CHEMBL1170349 | C34H58N12O8 | 762.914 | 11 / 12 | -5.2 | No |
| 135651 |  CHEMBL97420 CHEMBL97420 | C26H25Cl2N3 | 450.407 | 2 / 1 | 6.8 | No |
| 136286 |  CHEMBL1170035 CHEMBL1170035 | C35H58N10O7 | 730.912 | 10 / 9 | -4.0 | No |
| 137496 |  CHEMBL390834 CHEMBL390834 | C17H19ClN4O3S | 394.874 | 5 / 4 | 2.3 | Yes |
| 525510 | 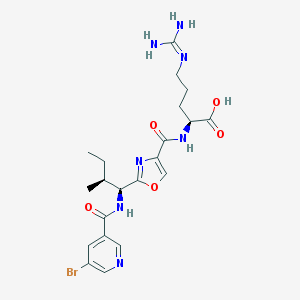 CHEMBL3735507 CHEMBL3735507 | C21H28BrN7O5 | 538.403 | 8 / 5 | 0.9 | No |
| 142679 | 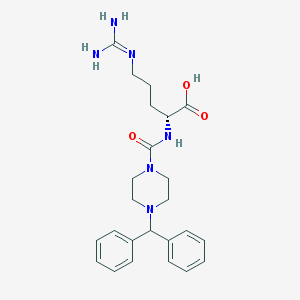 CHEMBL228900 CHEMBL228900 | C24H32N6O3 | 452.559 | 5 / 4 | -0.9 | Yes |
| 525673 | 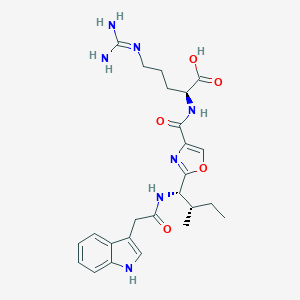 CHEMBL3735993 CHEMBL3735993 | C25H33N7O5 | 511.583 | 7 / 6 | 1.3 | No |
| 145719 | 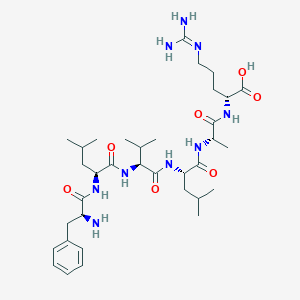 CHEMBL2021602 CHEMBL2021602 | C35H59N9O7 | 717.913 | 9 / 9 | -1.5 | No |
| 525760 |  CHEMBL3735568 CHEMBL3735568 | C25H32N6O5S | 528.628 | 8 / 5 | 3.0 | No |
| 149636 |  CID 49799040 CID 49799040 | C86H128N22O21 | 1806.1 | 24 / 26 | -4.5 | No |
| 149851 |  CHEMBL234521 CHEMBL234521 | C28H37N3O3 | 463.622 | 4 / 1 | 5.1 | No |
| 152771 |  CHEMBL1170044 CHEMBL1170044 | C34H54N10O8 | 730.868 | 10 / 9 | -4.9 | No |
| 158937 | 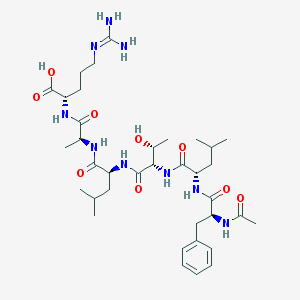 CHEMBL1170232 CHEMBL1170232 | C36H59N9O9 | 761.922 | 10 / 10 | 0.7 | No |
| 163967 | 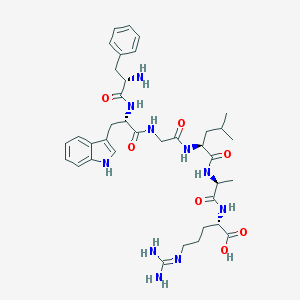 CHEMBL1170438 CHEMBL1170438 | C37H52N10O7 | 748.886 | 9 / 10 | -1.4 | No |
| 164264 | 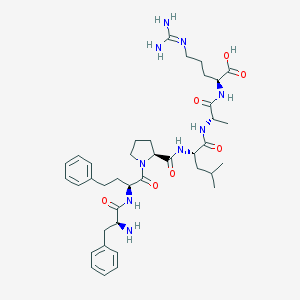 CHEMBL1170045 CHEMBL1170045 | C39H57N9O7 | 763.941 | 9 / 8 | -1.8 | No |
| 166549 | 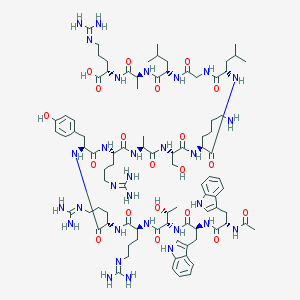 BDBM50388997 BDBM50388997 | C90H140N30O20 | 1962.3 | 25 / 30 | -6.1 | No |
| 168855 | 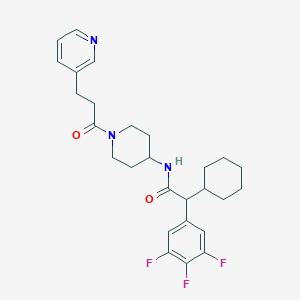 CHEMBL396030 CHEMBL396030 | C27H32F3N3O2 | 487.567 | 6 / 1 | 5.1 | No |
| 169179 | 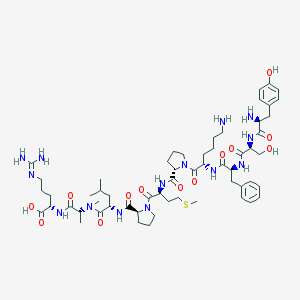 CHEMBL1170026 CHEMBL1170026 | C58H90N14O13S | 1223.5 | 17 / 13 | -2.2 | No |
| 175880 | 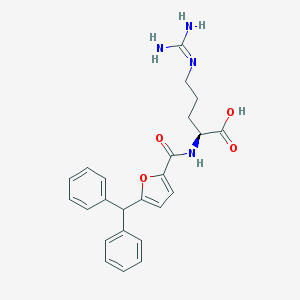 CHEMBL230034 CHEMBL230034 | C24H26N4O4 | 434.496 | 5 / 4 | 2.8 | Yes |
| 177389 | 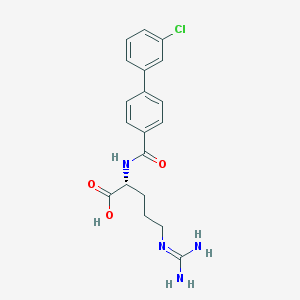 CHEMBL228846 CHEMBL228846 | C19H21ClN4O3 | 388.852 | 4 / 4 | 2.3 | Yes |
| 177506 | 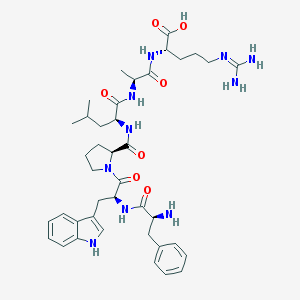 CHEMBL1170046 CHEMBL1170046 | C40H56N10O7 | 788.951 | 9 / 9 | -0.9 | No |
| 179027 | 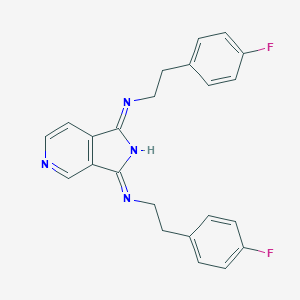 CHEMBL319113 CHEMBL319113 | C23H20F2N4 | 390.438 | 5 / 1 | 4.0 | Yes |
| 180703 | 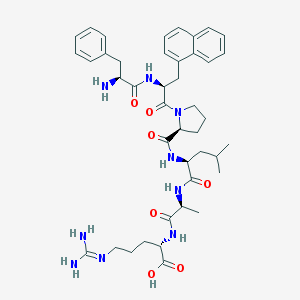 CHEMBL1170220 CHEMBL1170220 | C42H57N9O7 | 799.974 | 9 / 8 | 0.6 | No |
| 184110 | 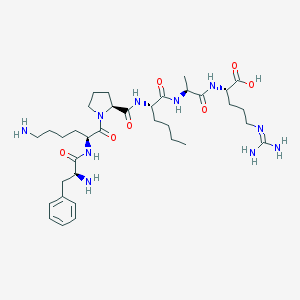 CHEMBL1170223 CHEMBL1170223 | C35H58N10O7 | 730.912 | 10 / 9 | -3.9 | No |
| 526798 | 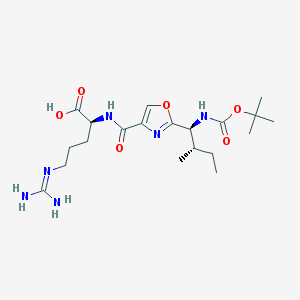 CHEMBL3736239 CHEMBL3736239 | C20H34N6O6 | 454.528 | 8 / 5 | 1.0 | Yes |
| 186775 | 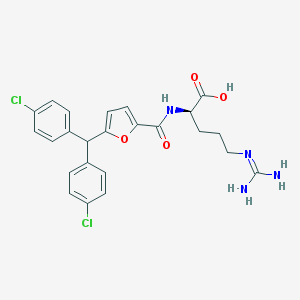 CHEMBL229006 CHEMBL229006 | C24H24Cl2N4O4 | 503.38 | 5 / 4 | 4.1 | No |
| 187703 | 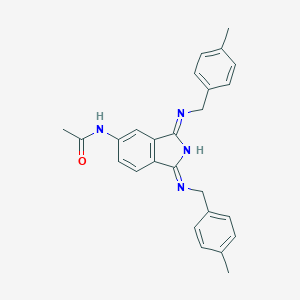 CHEMBL96138 CHEMBL96138 | C26H26N4O | 410.521 | 3 / 2 | 4.2 | Yes |
| 526893 | 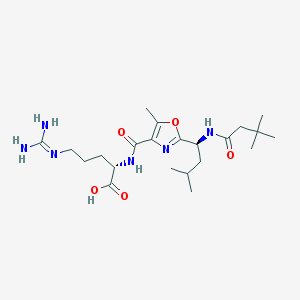 CHEMBL3734935 CHEMBL3734935 | C22H38N6O5 | 466.583 | 7 / 5 | 1.7 | Yes |
| 190553 |  CID 70684422 CID 70684422 | C92H139N31O20 | 1999.32 | 25 / 34 | -4.3 | No |
| 527081 |  CHEMBL3735614 CHEMBL3735614 | C22H27N3O5 | 413.474 | 5 / 4 | 1.9 | Yes |
| 195426 |  CHEMBL409773 CHEMBL409773 | C27H25Cl2N3O3 | 510.415 | 4 / 1 | 4.6 | No |
| 527279 |  CHEMBL3735180 CHEMBL3735180 | C26H38N6O7 | 546.625 | 9 / 5 | 1.6 | No |
| 203013 | 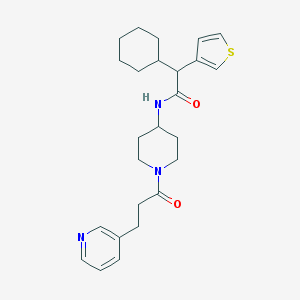 CHEMBL234005 CHEMBL234005 | C25H33N3O2S | 439.618 | 4 / 1 | 4.5 | Yes |
| 205123 | 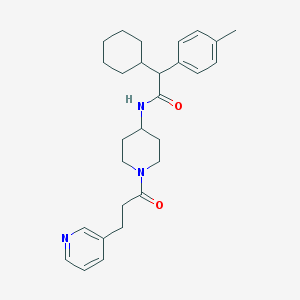 CHEMBL388204 CHEMBL388204 | C28H37N3O2 | 447.623 | 3 / 1 | 5.1 | No |
| 207639 | 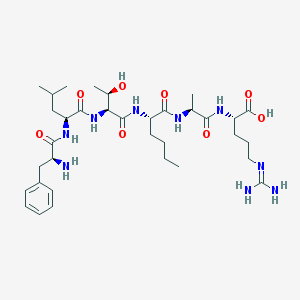 CHEMBL1170643 CHEMBL1170643 | C34H57N9O8 | 719.885 | 10 / 10 | -2.6 | No |
| 210230 | 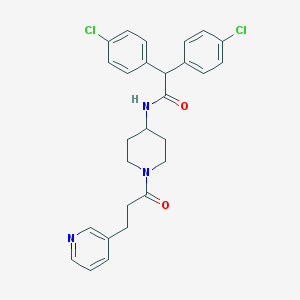 CHEMBL232418 CHEMBL232418 | C27H27Cl2N3O2 | 496.432 | 3 / 1 | 5.0 | Yes |
| 527556 | 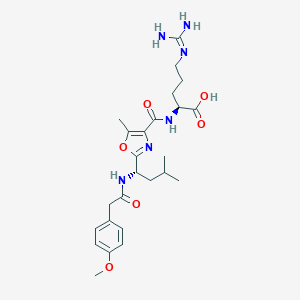 CHEMBL3735495 CHEMBL3735495 | C25H36N6O6 | 516.599 | 8 / 5 | 1.6 | No |
| 210785 | 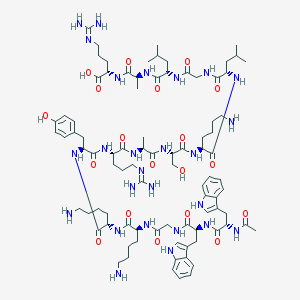 BDBM50388998 BDBM50388998 | C88H136N26O19 | 1862.22 | 24 / 27 | -3.9 | No |
| 527604 | 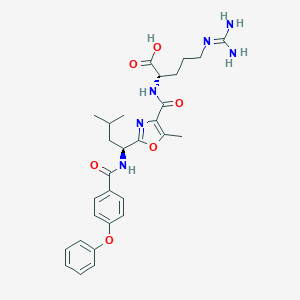 CHEMBL3736325 CHEMBL3736325 | C29H36N6O6 | 564.643 | 8 / 5 | 3.2 | No |
| 527634 | 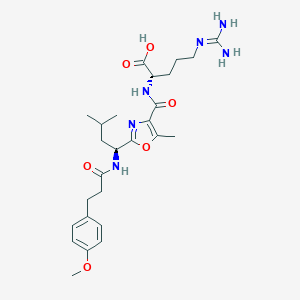 CHEMBL3735332 CHEMBL3735332 | C26H38N6O6 | 530.626 | 8 / 5 | 1.9 | No |
| 527667 | 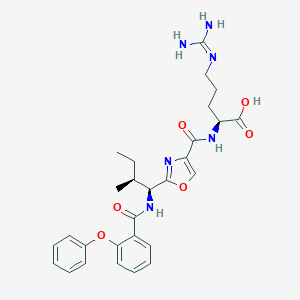 CHEMBL3735764 CHEMBL3735764 | C28H34N6O6 | 550.616 | 8 / 5 | 2.8 | No |
| 215618 | 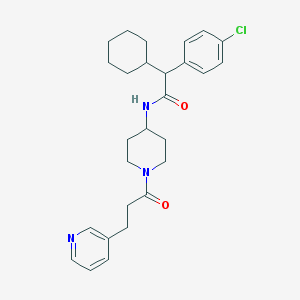 CHEMBL233675 CHEMBL233675 | C27H34ClN3O2 | 468.038 | 3 / 1 | 5.4 | No |
| 527861 | 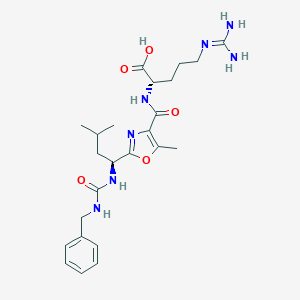 CHEMBL3734951 CHEMBL3734951 | C24H35N7O5 | 501.588 | 7 / 6 | 1.3 | No |
| 527868 | 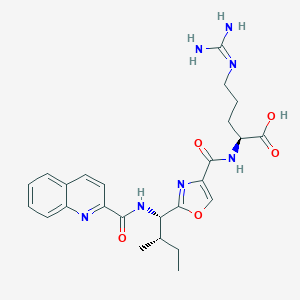 CHEMBL3735048 CHEMBL3735048 | C25H31N7O5 | 509.567 | 8 / 5 | 1.9 | No |
| 527923 | 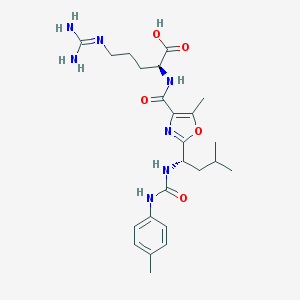 CHEMBL3736432 CHEMBL3736432 | C24H35N7O5 | 501.588 | 7 / 6 | 1.8 | No |
| 527980 | 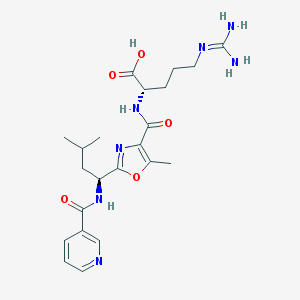 CHEMBL3735664 CHEMBL3735664 | C22H31N7O5 | 473.534 | 8 / 5 | 0.6 | Yes |
| 226514 | 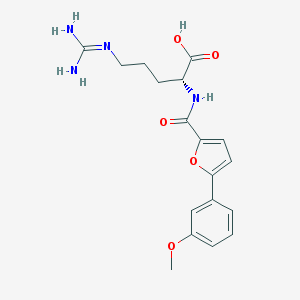 CHEMBL394538 CHEMBL394538 | C18H22N4O5 | 374.397 | 6 / 4 | 1.0 | Yes |
| 226665 | 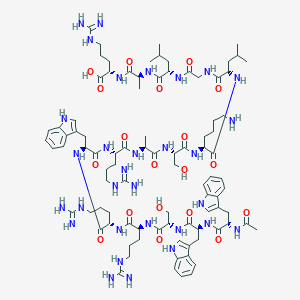 CID 70682339 CID 70682339 | C91H139N31O19 | 1971.31 | 24 / 34 | -4.5 | No |
| 230464 | 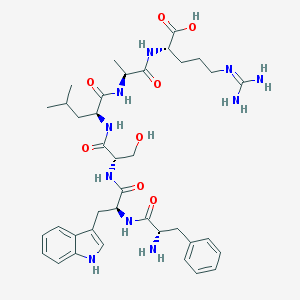 CHEMBL1170636 CHEMBL1170636 | C38H54N10O8 | 778.912 | 10 / 11 | -2.7 | No |
| 528198 | 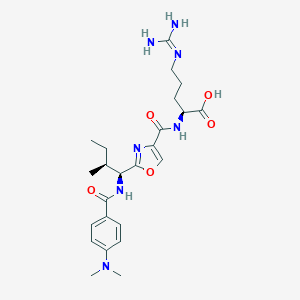 CHEMBL3736195 CHEMBL3736195 | C24H35N7O5 | 501.588 | 8 / 5 | 1.4 | No |
| 236881 | 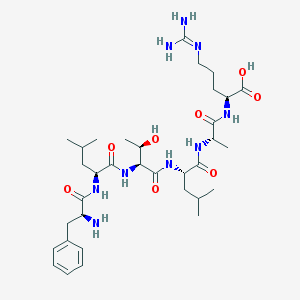 CHEMBL1170638 CHEMBL1170638 | C34H57N9O8 | 719.885 | 10 / 10 | -2.7 | No |
| 250944 | 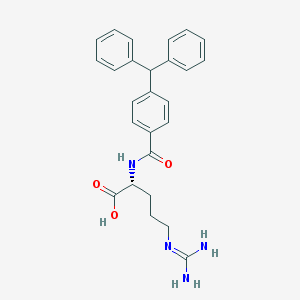 CHEMBL228901 CHEMBL228901 | C26H28N4O3 | 444.535 | 4 / 4 | 3.4 | Yes |
| 251229 | 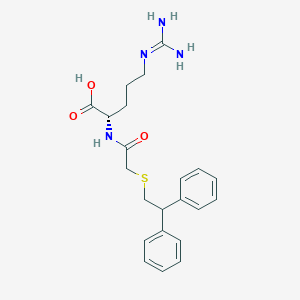 CHEMBL3342676 CHEMBL3342676 | C22H28N4O3S | 428.551 | 5 / 4 | 2.4 | Yes |
| 253297 | 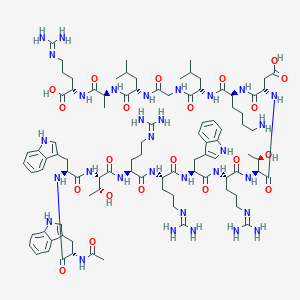 BDBM50389005 BDBM50389005 | C94H143N31O21 | 2043.37 | 26 / 31 | -6.0 | No |
| 254292 | 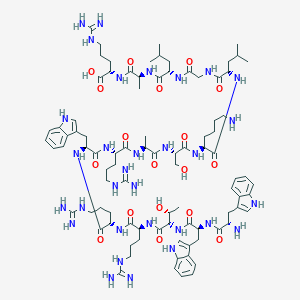 CID 70686567 CID 70686567 | C90H139N31O18 | 1943.3 | 24 / 34 | -4.3 | No |
| 451838 | 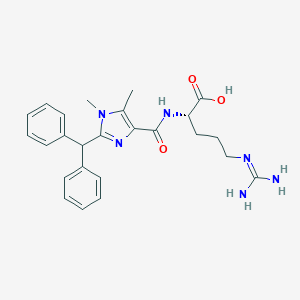 CHEMBL3342690 CHEMBL3342690 | C25H30N6O3 | 462.554 | 5 / 4 | 2.2 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417