

You can:
| Name | Melatonin receptor type 1B |
|---|---|
| Species | Gallus gallus (Chicken) |
| Gene | |
| Synonym | Mel-1B-R Mel1b receptor |
| Disease | N/A for non-human GPCRs |
| Length | 289 |
| Amino acid sequence | GNAFVVSLALADLVVALYPYPLVLLAIFHNGWTLGEMHCKVSGFVMGLSVIGSIFNITAIAINRYCYICHSFAYDKVYSCWNTMLYVSLIWVLTVIATVPNFFVGSLKYDPRIYSCTFVQTASSYYTIAVVVIHFIVPITVVSFCYLRIWVLVLQVRRRVKSETKPRLKPSDFRNFLTMFVVFVIFAFCWAPLNFIGLAVAINPSEMAPKVPEWLFIISYFMAYFNSCLNAIIYGLLNQNFRNEYKRILMSLWMPRLFFQDTSKGGTDGQKSKPSPALNNNDQMKTDTL |
| UniProt | P51050 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4466 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 25608 | 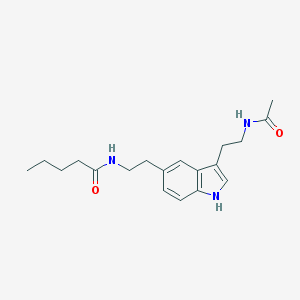 5-MeOT-N-pentanyl 5-MeOT-N-pentanyl | C19H27N3O2 | 329.444 | 2 / 3 | 2.6 | Yes |
| 34526 | 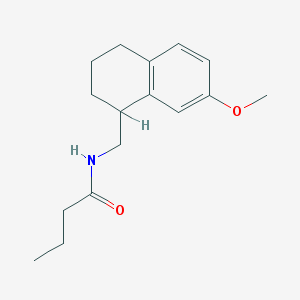 CHEMBL111866 CHEMBL111866 | C16H23NO2 | 261.365 | 2 / 1 | 3.0 | Yes |
| 67094 | 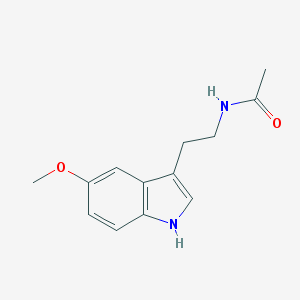 Melatonin Melatonin | C13H16N2O2 | 232.283 | 2 / 2 | 0.8 | Yes |
| 79523 | 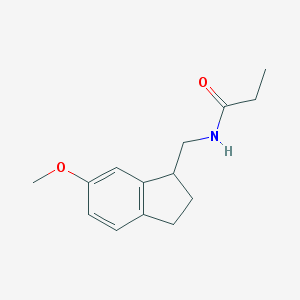 CHEMBL113244 CHEMBL113244 | C14H19NO2 | 233.311 | 2 / 1 | 2.1 | Yes |
| 92599 | 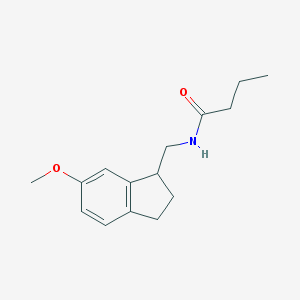 CHEMBL113243 CHEMBL113243 | C15H21NO2 | 247.338 | 2 / 1 | 2.5 | Yes |
| 112133 | 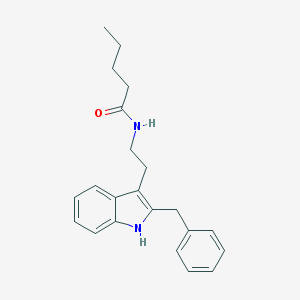 DH 97 DH 97 | C22H26N2O | 334.463 | 1 / 2 | 4.9 | Yes |
| 134197 | 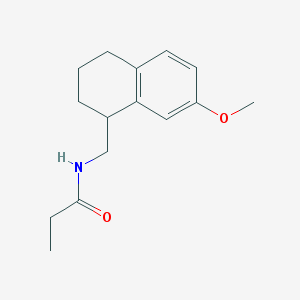 CHEMBL113859 CHEMBL113859 | C15H21NO2 | 247.338 | 2 / 1 | 2.7 | Yes |
| 179303 | 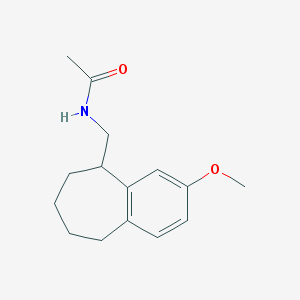 CHEMBL320136 CHEMBL320136 | C15H21NO2 | 247.338 | 2 / 1 | 2.7 | Yes |
| 182381 | 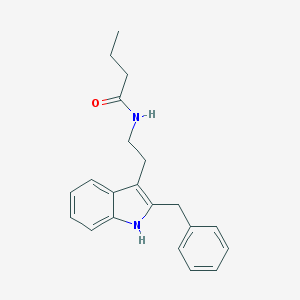 Luzindole,N-butanoyl Luzindole,N-butanoyl | C21H24N2O | 320.436 | 1 / 2 | 4.4 | Yes |
| 205646 | 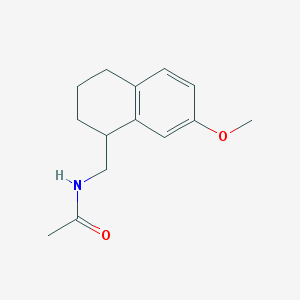 CHEMBL113067 CHEMBL113067 | C14H19NO2 | 233.311 | 2 / 1 | 2.2 | Yes |
| 208123 | 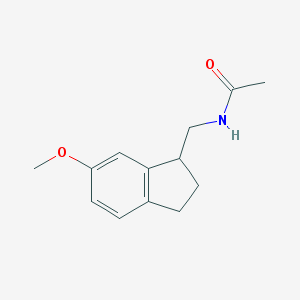 CHEMBL111667 CHEMBL111667 | C13H17NO2 | 219.284 | 2 / 1 | 1.7 | Yes |
| 210084 | 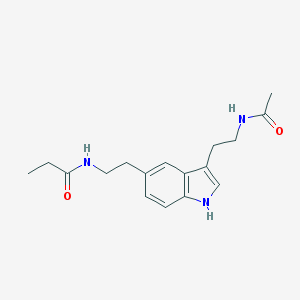 5-MeOT-N-propionyl 5-MeOT-N-propionyl | C17H23N3O2 | 301.39 | 2 / 3 | 1.7 | Yes |
| 244142 | 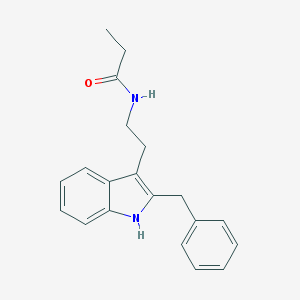 N-0889 N-0889 | C20H22N2O | 306.409 | 1 / 2 | 4.1 | Yes |
| 247924 | 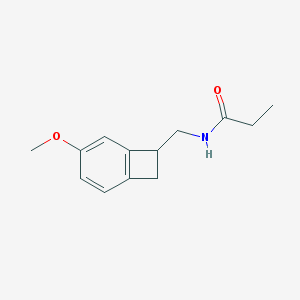 CHEMBL114929 CHEMBL114929 | C13H17NO2 | 219.284 | 2 / 1 | 1.0 | Yes |
| 265120 | 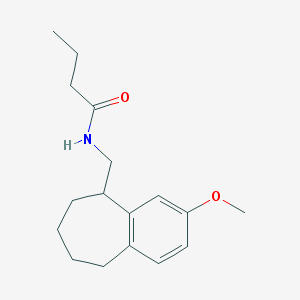 CHEMBL111879 CHEMBL111879 | C17H25NO2 | 275.392 | 2 / 1 | 3.6 | Yes |
| 280246 | 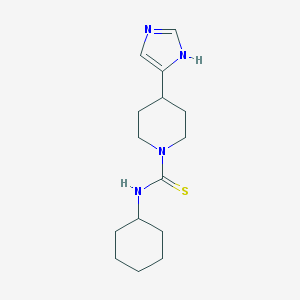 Thioperamide Thioperamide | C15H24N4S | 292.445 | 2 / 2 | 2.4 | Yes |
| 283255 | 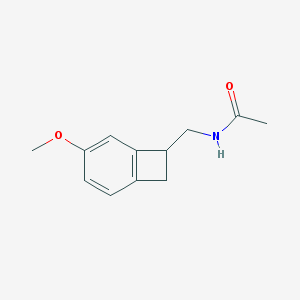 CHEMBL419939 CHEMBL419939 | C12H15NO2 | 205.257 | 2 / 1 | 0.6 | Yes |
| 288510 | 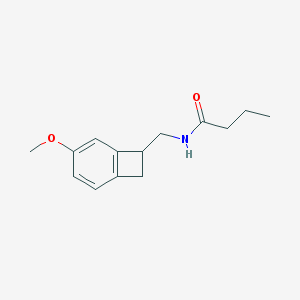 CHEMBL112164 CHEMBL112164 | C14H19NO2 | 233.311 | 2 / 1 | 1.4 | Yes |
| 290394 | 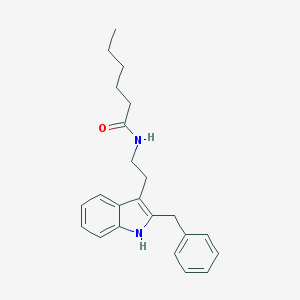 Luzindole,N-hexanoyl Luzindole,N-hexanoyl | C23H28N2O | 348.49 | 1 / 2 | 5.5 | No |
| 304736 | 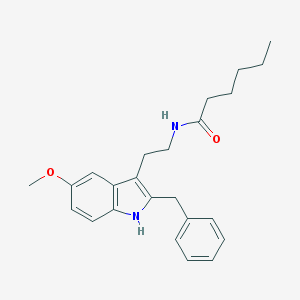 Luzindole,5-MeOT-N-hexanyl Luzindole,5-MeOT-N-hexanyl | C24H30N2O2 | 378.516 | 2 / 2 | 5.5 | No |
| 311648 | 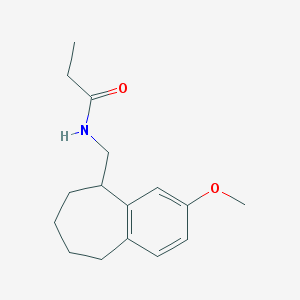 CHEMBL111880 CHEMBL111880 | C16H23NO2 | 261.365 | 2 / 1 | 3.2 | Yes |
| 332563 | 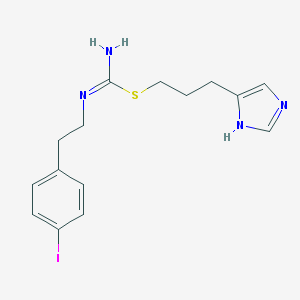 UNII-3SH45L9H3Z UNII-3SH45L9H3Z | C15H19IN4S | 414.309 | 3 / 2 | 3.3 | Yes |
| 352291 | 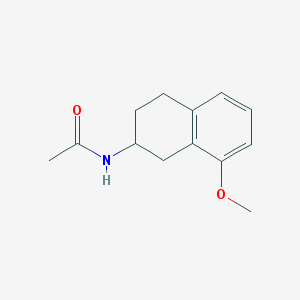 AH-001 AH-001 | C13H17NO2 | 219.284 | 2 / 1 | 1.9 | Yes |
| 375814 | 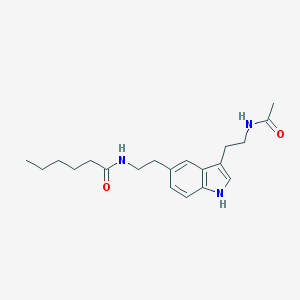 5-MeOT-N-hexanyl 5-MeOT-N-hexanyl | C20H29N3O2 | 343.471 | 2 / 3 | 3.1 | Yes |
| 383131 | 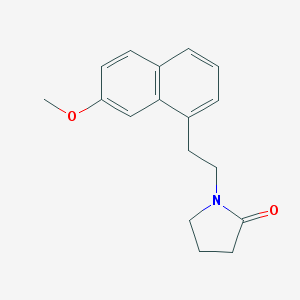 N-(2-(7-Methoxynaphth-1-yl)ethyl)pyrrolidin-2-one N-(2-(7-Methoxynaphth-1-yl)ethyl)pyrrolidin-2-one | C17H19NO2 | 269.344 | 2 / 0 | 3.0 | Yes |
| 383595 | 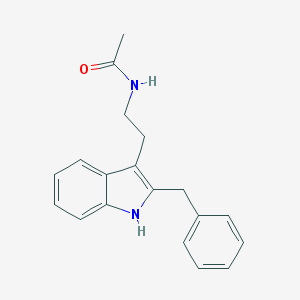 luzindole luzindole | C19H20N2O | 292.382 | 1 / 2 | 3.6 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417