

You can:
| Name | Alpha-2C adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRA2C |
| Synonym | alpha2-C4 Adra-2c ADRA2L2 ADRA2RL2 Adrenergic alpha2C- receptor class I [ Show all ] |
| Disease | Diabetic nephropathy; Fibromyalgia Hypertension Heart failure Glaucoma Alzheimer disease [ Show all ] |
| Length | 462 |
| Amino acid sequence | MASPALAAALAVAAAAGPNASGAGERGSGGVANASGASWGPPRGQYSAGAVAGLAAVVGFLIVFTVVGNVLVVIAVLTSRALRAPQNLFLVSLASADILVATLVMPFSLANELMAYWYFGQVWCGVYLALDVLFCTSSIVHLCAISLDRYWSVTQAVEYNLKRTPRRVKATIVAVWLISAVISFPPLVSLYRQPDGAAYPQCGLNDETWYILSSCIGSFFAPCLIMGLVYARIYRVAKLRTRTLSEKRAPVGPDGASPTTENGLGAAAGAGENGHCAPPPADVEPDESSAAAERRRRRGALRRGGRRRAGAEGGAGGADGQGAGPGAAESGALTASRSPGPGGRLSRASSRSVEFFLSRRRRARSSVCRRKVAQAREKRFTFVLAVVMGVFVLCWFPFFFSYSLYGICREACQVPGPLFKFFFWIGYCNSSLNPVIYTVFNQDFRRSFKHILFRRRRRGFRQ |
| UniProt | P18825 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P18825 |
| 3D structure model | This predicted structure model is from GPCR-EXP P18825. |
| BioLiP | N/A |
| Therapeutic Target Database | T01777 |
| ChEMBL | CHEMBL1916 |
| IUPHAR | 27 |
| DrugBank | BE0004864, BE0000342, BE0004888 |
| Name | brimonidine |
|---|---|
| Molecular formula | C11H10BrN5 |
| IUPAC name | 5-bromo-N-(4,5-dihydro-1H-imidazol-2-yl)quinoxalin-6-amine |
| Molecular weight | 292.14 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 0.6 |
| Synonyms | UK14304 CHEBI:3175 TC-148557 D03ACG UK 14304 [ Show all ] |
| Inchi Key | XYLJNLCSTIOKRM-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C11H10BrN5/c12-9-7(17-11-15-5-6-16-11)1-2-8-10(9)14-4-3-13-8/h1-4H,5-6H2,(H2,15,16,17) |
| PubChem CID | 2435 |
| ChEMBL | CHEMBL844 |
| IUPHAR | 520 |
| BindingDB | 34572 |
| DrugBank | DB00484 |
Structure | 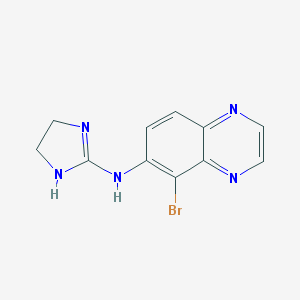 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| EC50 | 3.4 nM | PMID9016324 | BindingDB,ChEMBL |
| EC50 | 8.0 nM | None | ChEMBL |
| EC50 | 9.5 nM | N/A | BindingDB |
| EC50 | 9.55 nM | Bioorg. Med. Chem. Lett., (1995) 5:19:2255 | ChEMBL |
| Emax | 93.0 % | None | ChEMBL |
| IA | 0.82 - | PMID9016324 | ChEMBL |
| Intrinsic activity | 1.0 - | Bioorg. Med. Chem. Lett., (1995) 5:19:2255 | ChEMBL |
| Ki | 25.1189 - 1995.26 nM | PMID9824686, PMID9605427, PMID9760042, PMID9227000 | IUPHAR |
| Ki | 44.0 nM | PMID9016324 | BindingDB,ChEMBL |
| Ki | 83.0 nM | N/A | BindingDB |
| Ki | 83.18 nM | Bioorg. Med. Chem. Lett., (1995) 5:19:2255 | ChEMBL |
| Ki | 1949.84 nM | PMID9605427 | BindingDB |
| Ki | 1949.84 nM | PMID9605427 | PDSP |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417