

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MKALIFAAAGLLLLLPTFCQSGMENDTNNLAKPTLPIKTFRGAPPNSFEEFPFSALEGWTGATITVKIKCPEESASHLHVKNATMGYLTSSLSTKLIPAIYLLVFVVGVPANAVTLWMLFFRTRSICTTVFYTNLAIADFLFCVTLPFKIAYHLNGNNWVFGEVLCRATTVIFYGNMYCSILLLACISINRYLAIVHPFTYRGLPKHTYALVTCGLVWATVFLYMLPFFILKQEYYLVQPDITTCHDVHNTCESSSPFQLYYFISLAFFGFLIPFVLIIYCYAAIIRTLNAYDHRWLWYVKASLLILVIFTICFAPSNIILIIHHANYYYNNTDGLYFIYLIALCLGSLNSCLDPFLYFLMSKTRNHSTAYLTK | |
| CCCHHHHHCCHHHCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHCC | |
| 90048735120410543358888830103578999875667899987677997445676677777788898788886788986445664216885189999999999999999988986631677789999999999999999968999999847998988866799999999999999999999999984687640020244777402556779999999999989999722156068985799603897440179999999999999999999999999999999970577768738999999999999997899999999998741451479999999999999999988679674248418899997049 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MKALIFAAAGLLLLLPTFCQSGMENDTNNLAKPTLPIKTFRGAPPNSFEEFPFSALEGWTGATITVKIKCPEESASHLHVKNATMGYLTSSLSTKLIPAIYLLVFVVGVPANAVTLWMLFFRTRSICTTVFYTNLAIADFLFCVTLPFKIAYHLNGNNWVFGEVLCRATTVIFYGNMYCSILLLACISINRYLAIVHPFTYRGLPKHTYALVTCGLVWATVFLYMLPFFILKQEYYLVQPDITTCHDVHNTCESSSPFQLYYFISLAFFGFLIPFVLIIYCYAAIIRTLNAYDHRWLWYVKASLLILVIFTICFAPSNIILIIHHANYYYNNTDGLYFIYLIALCLGSLNSCLDPFLYFLMSKTRNHSTAYLTK | |
| 32000100011000010003343454454344442314114344443234122432443434423373423434344242444134324440210000210120023033232100000001333210000000000021000000001000003443111020000000011121010000000000100010000002023223221000000001100110110100012034277432010002134743322010001121112012201200220000001102346542100000000001000001231010000200231052240020011001000020002000000000440141033027 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHCCHHHCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHCC MKALIFAAAGLLLLLPTFCQSGMENDTNNLAKPTLPIKTFRGAPPNSFEEFPFSALEGWTGATITVKIKCPEESASHLHVKNATMGYLTSSLSTKLIPAIYLLVFVVGVPANAVTLWMLFFRTRSICTTVFYTNLAIADFLFCVTLPFKIAYHLNGNNWVFGEVLCRATTVIFYGNMYCSILLLACISINRYLAIVHPFTYRGLPKHTYALVTCGLVWATVFLYMLPFFILKQEYYLVQPDITTCHDVHNTCESSSPFQLYYFISLAFFGFLIPFVLIIYCYAAIIRTLNAYDHRWLWYVKASLLILVIFTICFAPSNIILIIHHANYYYNNTDGLYFIYLIALCLGSLNSCLDPFLYFLMSKTRNHSTAYLTK | |||||||||||||||||||||||||
| 1 | 5unfA | 0.22 | 0.26 | 0.97 | 3.30 | Download | LKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRH-GFDILVGQIDDAL----KLANEGKVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCQKGKKVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFPWQ-------ASYIVPLVWCMACLSSLPTFYFRDVRTIEYLGVNACIMAFP-PEKYAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMCEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR | |||||||||||||||||||
| 2 | 5vblA | 0.27 | 0.23 | 0.76 | 4.06 | Download | ----------------------------------------------------------------------------------------DWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSSRRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLRLRVSGAVATAVLWVLAALLAMPVMVLRTTGDLENTNKVQCYMDYSMVSSEWAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSPCDFDLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLL | |||||||||||||||||||
| 3 | 5unfA | 0.21 | 0.26 | 0.97 | 3.73 | Download | LKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEG-----KVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCQKPKKVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFP-------WQASYIVPLVWCMACLSSLPTFYFRDVRTIEYLGVNACIMAFPP-EKYAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMCEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR | |||||||||||||||||||
| 4 | 5nddA | 0.36 | 0.31 | 0.79 | 3.85 | Download | ---------------------------------------------------------------------------EFFSVDEFSASVLTGKLTTVFLPIVYTIVFVVALPSNGMALWVFLFRTKKAPAVIYMANLALADLLSVIWFPLKIAYHIHGNNWIYGEALCNVLIGFFYANMYCSILFLTCLSVQRAWEIVNPMGHSR-KKANIAIGISLAIWLLILLVTIPLYVVKQTIFIPALQITTCHDVLP-EQLLVGDMFNYFLSLAIGVFLFPAFLTASAYVLMIRAL-NSEKKRKRAIKLAVTVAAMYLICFTPSNLLLVVHYFLIKSQGQSHVYALYIVALCLSTLNSCIDPFVYYFVSHDFRDHAKNAL- | |||||||||||||||||||
| 5 | 5unfA | 0.22 | 0.26 | 0.97 | 3.68 | Download | LKVIEKADNAAQVKDALTKMRAAALDAQKATPPKL-----EDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCQKGKKVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFPW-------QASYIVPLVWCMACLSSLPTFYFRDVRTIEYLGVNACIMAFPP-EKYAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNSTRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMCEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR | |||||||||||||||||||
| 6 | 4djh | 0.26 | 0.23 | 0.74 | 1.54 | Download | -----------------------------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDD-YSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF | |||||||||||||||||||
| 7 | 4rwaA | 0.21 | 0.27 | 0.98 | 4.34 | Download | NAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQTTRNAYIQKYLGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSP--SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLC- | |||||||||||||||||||
| 8 | 4djh | 0.27 | 0.23 | 0.74 | 1.18 | Download | -----------------------------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDD-YSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF | |||||||||||||||||||
| 9 | 5unfA | 0.21 | 0.26 | 0.98 | 4.38 | Download | ADLEDNWETLNDNLKVIEKADNAAQRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCKGPKKVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFPW-------QASYIVPLVWCMACLSSLPTFYFRDVRTIEYLGVNACIMAFP-PEKYAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNSYGDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMGVIIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR | |||||||||||||||||||
| 10 | 4djh | 0.27 | 0.23 | 0.73 | 1.72 | Download | --------------------------------------------------------------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDD-DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFC- | |||||||||||||||||||
| ||||||||||||||||||||||||||
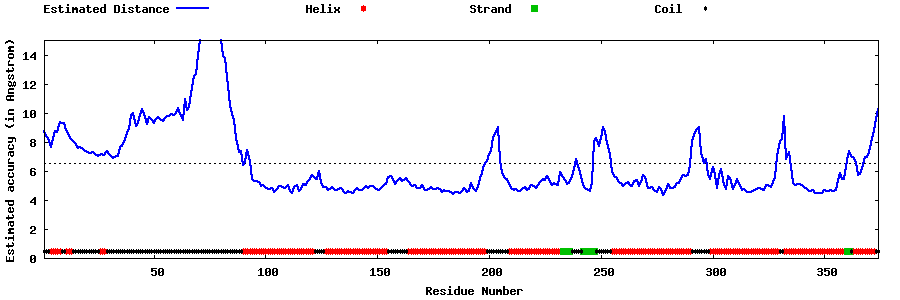
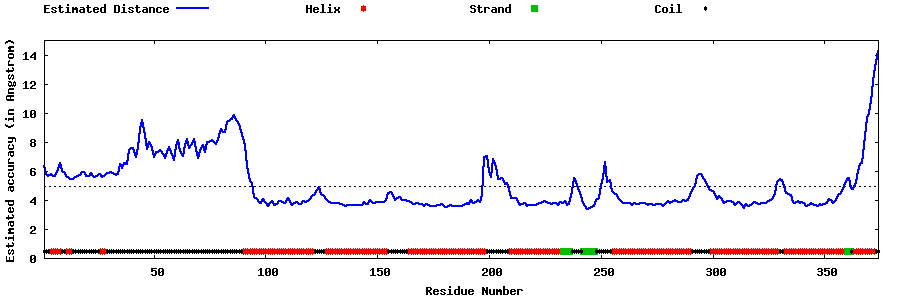
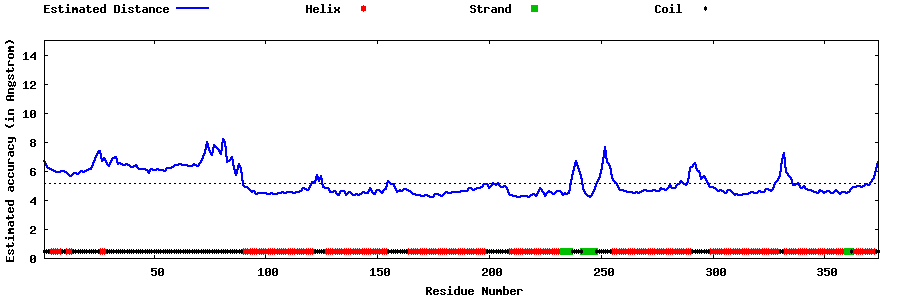
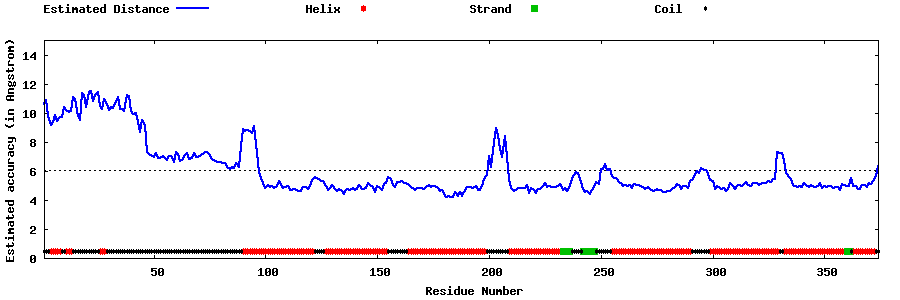
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||