

You can:
| Name | Mu-type opioid receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | OPRM1 |
| Synonym | hMOP M-OR-1 MOP opioid receptor, mu 1 opioid receptor [ Show all ] |
| Disease | Diarrhea Inflammatory disease Pain Major depressive disorder Migraine [ Show all ] |
| Length | 400 |
| Amino acid sequence | MDSSAAPTNASNCTDALAYSSCSPAPSPGSWVNLSHLDGNLSDPCGPNRTDLGGRDSLCPPTGSPSMITAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIINVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALVTIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCFREFCIPTSSNIEQQNSTRIRQNTRDHPSTANTVDRTNHQLENLEAETAPLP |
| UniProt | P35372 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P35372 |
| 3D structure model | This predicted structure model is from GPCR-EXP P35372. |
| BioLiP | N/A |
| Therapeutic Target Database | T47768 |
| ChEMBL | CHEMBL233 |
| IUPHAR | 319 |
| DrugBank | BE0000770 |
| Name | morphine |
|---|---|
| Molecular formula | C17H19NO3 |
| IUPAC name | (4R,4aR,7S,7aR,12bS)-3-methyl-2,4,4a,7,7a,13-hexahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline-7,9-diol |
| Molecular weight | 285.343 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 2 |
| XlogP | 0.8 |
| Synonyms | Morphine;4-methyl-(1S,5R,13R,14S,17R)-12-oxa-4-azapentacyclo[9.6.1.01,13.05,17.07,18]octadeca-7(18),8,10,15-tetraene-10,14-diol Aguettant Ms Emma BQJCRHHNABKAKU-KBQPJGBKSA-N Oramorph [ Show all ] |
| Inchi Key | BQJCRHHNABKAKU-KBQPJGBKSA-N |
| Inchi ID | InChI=1S/C17H19NO3/c1-18-7-6-17-10-3-5-13(20)16(17)21-15-12(19)4-2-9(14(15)17)8-11(10)18/h2-5,10-11,13,16,19-20H,6-8H2,1H3/t10-,11+,13-,16-,17-/m0/s1 |
| PubChem CID | 5288826 |
| ChEMBL | CHEMBL70 |
| IUPHAR | 1627 |
| BindingDB | 50000092 |
| DrugBank | DB00295 |
Structure | 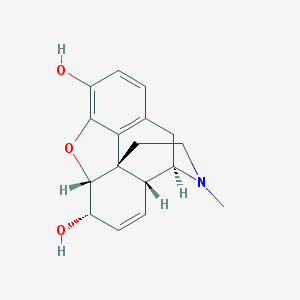 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Activity | 93.0 % | PMID16913723, PMID19253983, PMID17887741 | ChEMBL |
| Activity | 103.0 % | PMID21235243 | ChEMBL |
| EC50 | 3.7 nM | PMID24900459 | BindingDB,ChEMBL |
| EC50 | 6.0 nM | PMID17616524 | ChEMBL |
| EC50 | 6.4 nM | PMID17616524 | ChEMBL |
| EC50 | 7.0 nM | PMID17616524 | ChEMBL |
| EC50 | 10.0 nM | PMID25087049 | BindingDB |
| EC50 | 10.0 nM | PMID25087049 | ChEMBL |
| EC50 | 15.6 nM | PMID16913723, PMID19253983, PMID17887741 | ChEMBL |
| EC50 | 16.0 nM | PMID16913723, PMID19253983, PMID17887741 | BindingDB |
| EC50 | 34.0 nM | N/A | BindingDB |
| EC50 | 39.81 nM | PMID24063433 | ChEMBL |
| EC50 | 50.0 nM | PMID23880538 | ChEMBL |
| EC50 | 316.0 nM | PMID21235243, PMID17887741 | BindingDB,ChEMBL |
| EC50 | 501.19 nM | PMID24063433 | ChEMBL |
| ED50 | 37.0 nM | PMID17625813, PMID20055417 | ChEMBL |
| Emax | 77.0 % | PMID22995061 | ChEMBL |
| Emax | 86.0 % | PMID17625813, PMID20055417 | ChEMBL |
| Emax | 100.0 % | PMID17616524 | ChEMBL |
| Emax | 202.0 % | PMID18062664 | ChEMBL |
| IC50 | 0.57 nM | PMID12502358 | BindingDB |
| IC50 | 0.57 nM | PMID12502358 | ChEMBL |
| IC50 | 1.0 nM | PMID11585443 | BindingDB |
| IC50 | 2.5 nM | PMID24171469 | ChEMBL |
| IC50 | 4.3 nM | PMID22995061 | BindingDB,ChEMBL |
| IC50 | 6.3 nM | PMID26789491 | BindingDB,ChEMBL |
| Inhibition | 81.0 % | PMID24183585 | ChEMBL |
| Inhibition | 91.0 % | PMID23880538 | ChEMBL |
| Inhibition | 98.0 % | PMID24183585 | ChEMBL |
| Ki | 0.14 nM | PMID22680612 | BindingDB |
| Ki | 0.14 nM | PMID22680612 | ChEMBL |
| Ki | 0.32 nM | N/A | BindingDB |
| Ki | 0.53 nM | PMID26390077 | ChEMBL |
| Ki | 0.53 nM | PMID26390077 | BindingDB |
| Ki | 0.88 nM | PMID22439881, PMID16392810, PMID15055988 | ChEMBL |
| Ki | 0.88 nM | PMID22439881, PMID16392810, PMID15055988 | BindingDB |
| Ki | 1.0 nM | PMID9686407, PMID2549383 | IUPHAR |
| Ki | 1.02 nM | PMID7932177 | BindingDB |
| Ki | 1.1 nM | PMID16913723, PMID17887741, PMID9686407 | BindingDB,ChEMBL |
| Ki | 1.7 nM | PMID18311899 | ChEMBL |
| Ki | 1.8 nM | PMID2567782 | BindingDB,ChEMBL |
| Ki | 1.9 nM | PMID21621410 | BindingDB |
| Ki | 1.92 nM | PMID21621410 | ChEMBL |
| Ki | 2.0 nM | PMID7815359 | BindingDB |
| Ki | 2.2 nM | PMID24900459, PMID12513698 | BindingDB,ChEMBL |
| Ki | 2.5 nM | PMID21621410 | BindingDB |
| Ki | 2.52 nM | PMID21621410 | ChEMBL |
| Ki | 2.55 nM | PMID19627147, PMID21684752 | ChEMBL |
| Ki | 2.6 nM | PMID19627147, PMID21684752, PMID23880358, PMID21570305, PMID25599950, PMID23618710, PMID22341895 | BindingDB,ChEMBL |
| Ki | 3.0 nM | PMID12930147 | BindingDB,ChEMBL |
| Ki | 3.96 nM | PMID12372519 | ChEMBL |
| Ki | 4.0 nM | PMID12372519, PMID17616524 | BindingDB,ChEMBL |
| Ki | 4.6 nM | PMID21621410 | BindingDB,ChEMBL |
| Ki | 5.2 nM | PMID25062506, MedChemComm, (2016) 7:2:317 | BindingDB,ChEMBL |
| Ki | 14.0 nM | PMID8114680 | BindingDB |
| Ki | 17.0 nM | PMID12166947, PMID9651168 | BindingDB,ChEMBL |
| Ki | 22.0 nM | PMID25494650 | BindingDB,ChEMBL |
| Ki | 38.0 nM | , Bioorg. Med. Chem. Lett., (1992) 2:7:715 | BindingDB,ChEMBL |
| Log10ED50 | 7.13 - | PMID18062664 | ChEMBL |
| Ratio | 0.021 - | PMID2567782 | ChEMBL |
| Ratio | 265.0 - | PMID12502358 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417