

You can:
| Name | Hydroxycarboxylic acid receptor 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HCAR2 |
| Synonym | G protein-coupled receptor 109A PUMAG Nicotinic acid receptor Nic1 Niacr1 [ Show all ] |
| Disease | Type 2 diabetes Hyperlipidaemia Major depressive disorder Cardiovascular disorder Atherosclerosis [ Show all ] |
| Length | 363 |
| Amino acid sequence | MNRHHLQDHFLEIDKKNCCVFRDDFIVKVLPPVLGLEFIFGLLGNGLALWIFCFHLKSWKSSRIFLFNLAVADFLLIICLPFLMDNYVRRWDWKFGDIPCRLMLFMLAMNRQGSIIFLTVVAVDRYFRVVHPHHALNKISNRTAAIISCLLWGITIGLTVHLLKKKMPIQNGGANLCSSFSICHTFQWHEAMFLLEFFLPLGIILFCSARIIWSLRQRQMDRHAKIKRAITFIMVVAIVFVICFLPSVVVRIRIFWLLHTSGTQNCEVYRSVDLAFFITLSFTYMNSMLDPVVYYFSSPSFPNFFSTLINRCLQRKMTGEPDNNRSTSVELTGDPNKTRGAPEALMANSGEPWSPSYLGPTSP |
| UniProt | Q8TDS4 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q8TDS4 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q8TDS4. |
| BioLiP | N/A |
| Therapeutic Target Database | T00864 |
| ChEMBL | CHEMBL3785 |
| IUPHAR | 312 |
| DrugBank | BE0000635 |
| Name | nicotinic acid |
|---|---|
| Molecular formula | C6H5NO2 |
| IUPAC name | pyridine-3-carboxylic acid |
| Molecular weight | 123.111 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 0.4 |
| Synonyms | Nicotinic acid, SAJ special grade, >=99.5% Niacin (Nicotinic Acid), Pharmaceutical Secondary Standard; Certified Reference Material Niacinamide Astra Brand GTPL1588 Niaspan [ Show all ] |
| Inchi Key | PVNIIMVLHYAWGP-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C6H5NO2/c8-6(9)5-2-1-3-7-4-5/h1-4H,(H,8,9) |
| PubChem CID | 938 |
| ChEMBL | CHEMBL573 |
| IUPHAR | 1594, 1588 |
| BindingDB | 23515 |
| DrugBank | DB00627 |
Structure | 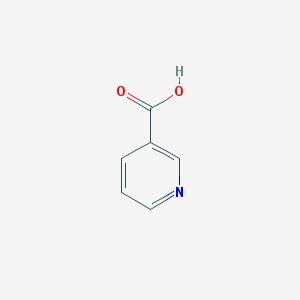 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Activity | 100.0 % | PMID18752940, PMID19592242 | ChEMBL |
| Activity | 200.0 % | PMID19524438 | ChEMBL |
| EC50 | <1000.0 nM | PMID17804224 | ChEMBL |
| EC50 | 8.7 nM | PMID17804224 | BindingDB |
| EC50 | 8.71 nM | PMID17804224 | ChEMBL |
| EC50 | 26.92 nM | PMID19524438 | ChEMBL |
| EC50 | 27.0 nM | PMID20363624, PMID19524438 | BindingDB,ChEMBL |
| EC50 | 47.0 nM | PMID25737085 | ChEMBL |
| EC50 | 51.0 nM | PMID22435740 | BindingDB,ChEMBL |
| EC50 | 63.1 - 1000.0 nM | PMID12522134, PMID12563315, PMID12646212 | IUPHAR |
| EC50 | 99.0 nM | PMID24900372 | BindingDB,ChEMBL |
| EC50 | 100.0 nM | PMID24900295, PMID22209457 | BindingDB,ChEMBL |
| EC50 | 120.0 nM | PMID17358052, PMID17588745 | BindingDB,ChEMBL |
| EC50 | 527.0 nM | PMID20184326 | BindingDB,ChEMBL |
| EC50 | 580.0 nM | PMID17452318 | BindingDB,ChEMBL |
| EC50 | 730.0 nM | PMID26784936 | BindingDB,ChEMBL |
| EC50 | 780.0 nM | PMID24900524 | BindingDB,ChEMBL |
| EC50 | 1000.0 nM | PMID17994679, PMID20444602, PMID19309152, PMID18029181, PMID18752940, PMID19592242, PMID20452209 | BindingDB,ChEMBL |
| EC50 | 1400.0 nM | PMID18760600 | BindingDB,ChEMBL |
| Efficacy | 95.0 % | PMID17804224, PMID22435740 | ChEMBL |
| Efficacy | 100.0 % | PMID20363624 | ChEMBL |
| Fold change | 0.0 - | PMID17994679 | ChEMBL |
| IC50 | 130.0 nM | PMID18760600 | BindingDB,ChEMBL |
| IC50 | 140.0 nM | PMID17994679, PMID20444602, PMID19307116, PMID20615702, PMID19309152, PMID18029181, PMID19592242, PMID20452209 | BindingDB,ChEMBL |
| IC50 | 150.0 nM | PMID18752940 | BindingDB,ChEMBL |
| IC50 | 249.0 nM | PMID25737085 | ChEMBL |
| IC50 | 67300.0 nM | PMID17452318 | BindingDB,ChEMBL |
| Inhibition | 100.0 % | PMID17452318 | ChEMBL |
| Kd | 50.1 - 100.0 nM | PMID12522134, PMID12563315, PMID12646212 | IUPHAR |
| Ki | 50.0 nM | PMID18665582 | BindingDB,ChEMBL |
| Ki | 82.0 nM | PMID20184326 | BindingDB,ChEMBL |
| Ki | 104.0 nM | PMID20184326 | BindingDB,ChEMBL |
| Ratio | 0.8 - | PMID25737085 | ChEMBL |
| Ratio IC50 | 0.0 - | PMID19309152 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417