

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNRHHLQDHFLEIDKKNCCVFRDDFIVKVLPPVLGLEFIFGLLGNGLALWIFCFHLKSWKSSRIFLFNLAVADFLLIICLPFLMDNYVRRWDWKFGDIPCRLMLFMLAMNRQGSIIFLTVVAVDRYFRVVHPHHALNKISNRTAAIISCLLWGITIGLTVHLLKKKMPIQNGGANLCSSFSICHTFQWHEAMFLLEFFLPLGIILFCSARIIWSLRQRQMDRHAKIKRAITFIMVVAIVFVICFLPSVVVRIRIFWLLHTSGTQNCEVYRSVDLAFFITLSFTYMNSMLDPVVYYFSSPSFPNFFSTLINRCLQRKMTGEPDNNRSTSVELTGDPNKTRGAPEALMANSGEPWSPSYLGPTSP | |
| CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 998755655667888766666532789749999999999999889984461025277883889999999999999999789999999579998777655999999999999999999999999724653231312114145788999999999999999889866458860890699825873078999989500179999999999999999981676766552116899999999999998467999999999998516789817999999999999999999999989999813988889999999987156666788888775545268999888786731378999999988999998 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNRHHLQDHFLEIDKKNCCVFRDDFIVKVLPPVLGLEFIFGLLGNGLALWIFCFHLKSWKSSRIFLFNLAVADFLLIICLPFLMDNYVRRWDWKFGDIPCRLMLFMLAMNRQGSIIFLTVVAVDRYFRVVHPHHALNKISNRTAAIISCLLWGITIGLTVHLLKKKMPIQNGGANLCSSFSICHTFQWHEAMFLLEFFLPLGIILFCSARIIWSLRQRQMDRHAKIKRAITFIMVVAIVFVICFLPSVVVRIRIFWLLHTSGTQNCEVYRSVDLAFFITLSFTYMNSMLDPVVYYFSSPSFPNFFSTLINRCLQRKMTGEPDNNRSTSVELTGDPNKTRGAPEALMANSGEPWSPSYLGPTSP | |
| 635443443234244431232445021000021012002301310120010001223433101000000000110000001010000035330200300120010111311110000000003100000010020232232210000000001000000001000120354742210101223432201001102113312210020001000101325366444220000000000000100132000000011002323234414123002001100100013000210200000046014102400330144334643543444345444454424334533346664333464263768 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MNRHHLQDHFLEIDKKNCCVFRDDFIVKVLPPVLGLEFIFGLLGNGLALWIFCFHLKSWKSSRIFLFNLAVADFLLIICLPFLMDNYVRRWDWKFGDIPCRLMLFMLAMNRQGSIIFLTVVAVDRYFRVVHPHHALNKISNRTAAIISCLLWGITIGLTVHLLKKKMPIQNGGANLCSSFSICHTFQWHEAMFLLEFFLPLGIILFCSARIIWSLRQRQMDRHAKIKRAITFIMVVAIVFVICFLPSVVVRIRIFWLLHTSGTQNCEVYRSVDLAFFITLSFTYMNSMLDPVVYYFSSPSFPNFFSTLINRCLQRKMTGEPDNNRSTSVELTGDPNKTRGAPEALMANSGEPWSPSYLGPTSP | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.23 | 0.20 | 0.80 | 3.30 | Download | ---------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK-EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEF-FGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------------------------------- | |||||||||||||||||||
| 2 | 4xnwA | 0.29 | 0.24 | 0.81 | 4.24 | Download | -------------SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVKNKTITCYDTTSDEYLRYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRALDFQTPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR-------------------------------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.23 | 0.20 | 0.80 | 3.69 | Download | ---------PCQKINV------KQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK-EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTF-QEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.26 | 0.24 | 0.76 | 1.54 | Download | ----------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYSFMICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.26 | 0.24 | 0.76 | 1.15 | Download | ----------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDDYFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEAL---GS---A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------------------- | |||||||||||||||||||
| 6 | 5o9hA | 0.23 | 0.22 | 0.80 | 3.34 | Download | ---------------------TLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAK-RTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEPPKVLCGVDHDRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSA---RETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFL-----EPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------------------------- | |||||||||||||||||||
| 7 | 5t1a | 0.24 | 0.25 | 0.78 | 1.70 | Download | -----------------------QIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKED-SVYVCGPYPRGWNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFG-LSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------------------------------------------ | |||||||||||||||||||
| 8 | 4mbsA | 0.22 | 0.20 | 0.78 | 4.58 | Download | ---------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFQTLKIVILGL----VLPLLVMVICYSGILKTLLREKKRHRD-----VRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFG-LNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------------------------------- | |||||||||||||||||||
| 9 | 4yayA | 0.23 | 0.20 | 0.79 | 3.53 | Download | LNSS-----------DCPKAGRHNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF--IITVCAFHYETLPIGLGLTKNILGFLFPFLIILTSYTLIWKAL--------KKNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQL-GIIRDCRIADIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------------------------------------------------------ | |||||||||||||||||||
| 10 | 5c1mA | 0.25 | 0.24 | 0.79 | 4.38 | Download | -----------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKLRRITRMVLVVVAVFIVCWTPIHIYVIIKAL-------ITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
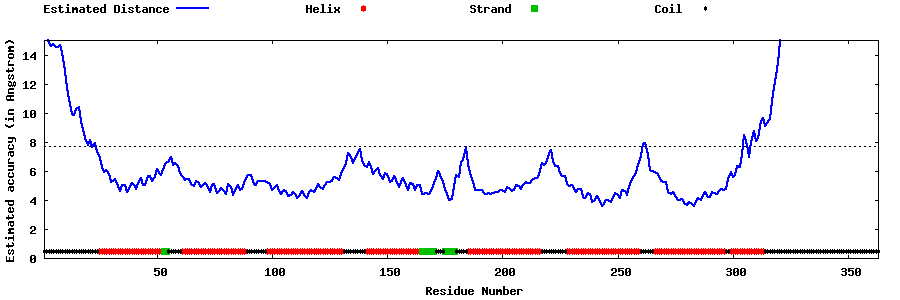 | |||
|
|
|||
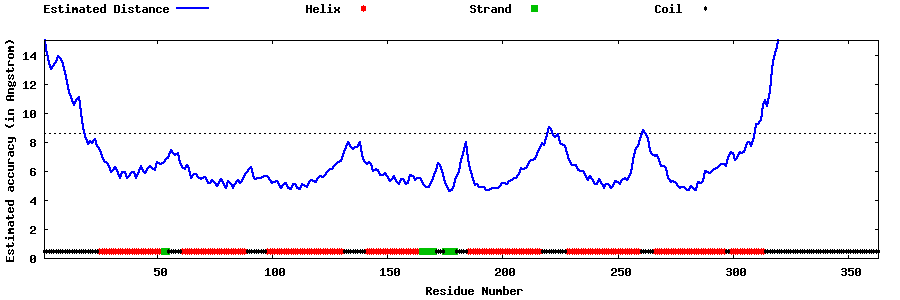 | |||
|
| |||
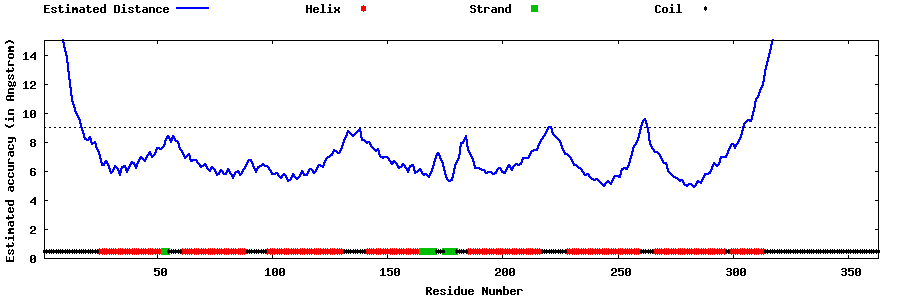 | |||
|
| |||
 | |||
|
| |||
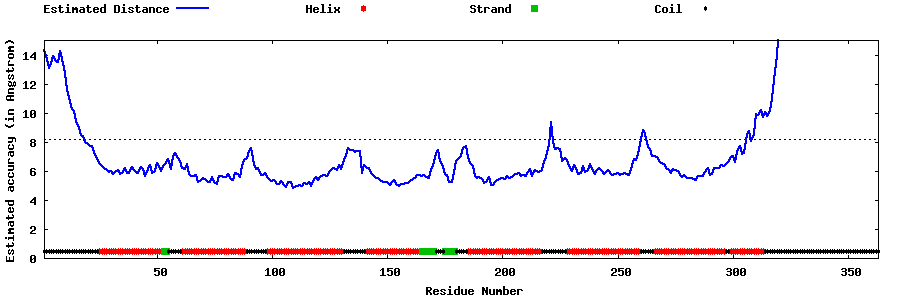 | |||
|
| |||