

You can:
| Name | 5-hydroxytryptamine receptor 1A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR1A |
| Synonym | 5-HT-1A 5-HT1A serotonin receptor 1A 5-HT1A receptor 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled [ Show all ] |
| Disease | Urinary incontinence Generalized anxiety disorder Generalized anxiety disorder; Social phobia Hypertension Hypoactive sexual desire disorder [ Show all ] |
| Length | 422 |
| Amino acid sequence | MDVLSPGQGNNTTSPPAPFETGGNTTGISDVTVSYQVITSLLLGTLIFCAVLGNACVVAAIALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPEDRSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVKKVEKTGADTRHGASPAPQPKKSVNGESGSRNWRLGVESKAGGALCANGAVRQGDDGAALEVIEVHRVGNSKEHLPLPSEAGPTPCAPASFERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHMPTLLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFCRQ |
| UniProt | P08908 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P08908 |
| 3D structure model | This predicted structure model is from GPCR-EXP P08908. |
| BioLiP | N/A |
| Therapeutic Target Database | T78709 |
| ChEMBL | CHEMBL214 |
| IUPHAR | 1 |
| DrugBank | BE0000291 |
| Name | Bmy-7378 |
|---|---|
| Molecular formula | C22H31N3O3 |
| IUPAC name | 8-[2-[4-(2-methoxyphenyl)piperazin-1-yl]ethyl]-8-azaspiro[4.5]decane-7,9-dione |
| Molecular weight | 385.508 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 0 |
| XlogP | 2.8 |
| Synonyms | 8-[2-[4-(2-methoxyphenyl)-1-piperazinyl]ethyl]-8-azaspiro[4.5]decane-7,9-dione AKOS030559908 CCG-204301 Lopac-B-134 NCGC00015126-05 [ Show all ] |
| Inchi Key | AYYCFGDXLUPJAQ-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C22H31N3O3/c1-28-19-7-3-2-6-18(19)24-13-10-23(11-14-24)12-15-25-20(26)16-22(17-21(25)27)8-4-5-9-22/h2-3,6-7H,4-5,8-17H2,1H3 |
| PubChem CID | 2419 |
| ChEMBL | CHEMBL13647 |
| IUPHAR | 9 |
| BindingDB | 50026917 |
| DrugBank | N/A |
Structure | 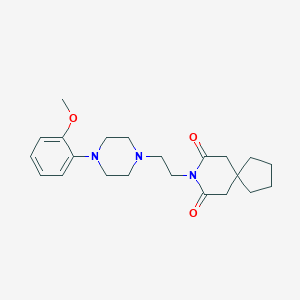 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| %max | 26.0 - | PMID18817363 | ChEMBL |
| Activity | 0.3 - | PMID15055991 | ChEMBL |
| Emax | 26.0 % | PMID23252794, PMID22145629, PMID27689727, PMID20185311 | ChEMBL |
| Emax | 26.0 - | PMID15686938 | ChEMBL |
| Emax | 100.0 % | PMID20605276 | ChEMBL |
| IC50 | 10.0 - 158.489 nM | PMID9048968 | IUPHAR |
| Ki | 0.37 nM | PMID23252794 | BindingDB |
| Ki | 0.3715 nM | PMID18817363, PMID11931617, PMID23252794 | ChEMBL |
| Ki | 0.372 nM | PMID18817363 | BindingDB |
| Ki | 0.46 nM | PMID15828846 | BindingDB,ChEMBL |
| Ki | 0.8 nM | PMID15055991 | BindingDB,ChEMBL |
| Ki | 0.9333 nM | PMID10514291 | ChEMBL |
| Ki | 1.259 nM | PMID20605276, PMID20185311, PMID15686938, PMID27689727, PMID22145629 | ChEMBL |
| Ki | 1.26 nM | PMID20605276, PMID22145629, PMID20185311 | BindingDB |
| Ki | 1.3 nM | PMID27689727 | BindingDB |
| Ki | 1.7 nM | PMID9871765 | BindingDB |
| Ki | 1.738 nM | PMID9822553 | ChEMBL |
| Ki ratio | 8.9 - | PMID7562940 | ChEMBL |
| Max | 26.0 % | PMID10514291 | ChEMBL |
| pD2 | 7.83 - | PMID20605276 | ChEMBL |
| pD2 | 9.27 - | PMID18817363, PMID15686938, PMID10514291, PMID22145629, PMID20185311, PMID23252794, PMID27689727 | ChEMBL |
| Ratio | 1.0 - | PMID15055991 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417