

You can:
| Name | 5-hydroxytryptamine receptor 2B |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR2B |
| Synonym | stomach fundus serotonin receptor serotonin receptor 2B 5-HT2F 5-HT-2B 5-HT2B [ Show all ] |
| Disease | Psychoses Migraine Irritable bowel syndrome Depression; Cerebral infarction Coronary heart disease [ Show all ] |
| Length | 481 |
| Amino acid sequence | MALSYRVSELQSTIPEHILQSTFVHVISSNWSGLQTESIPEEMKQIVEEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIMHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETDVDNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAYLVKNKPPQRLTWLTVSTVFQRDETPCSSPEKVAMLDGSRKDKALPNSGDETLMRRTSTIGKKSVQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYRATKSVKTLRKRSSKIYFRNPMAENSKFFKKHGIRNGINPAMYQSPMRLRSSTIQSSSIILLDTLLLTENEGDKTEEQVSYV |
| UniProt | P41595 |
| Protein Data Bank | 5tud, 5tvn, 6drx, 4ib4, 4nc3, 6dry, 6drz, 6ds0 |
| GPCR-HGmod model | P41595 |
| 3D structure model | This structure is from PDB ID 5tud. |
| BioLiP | BL0425280, BL0425283, BL0425282, BL0239858, BL0239859, BL0265182, BL0265183, BL0368464, BL0425281, BL0385686,BL0385687, BL0425284, BL0425285, BL0425286, BL0368465, BL0425287 |
| Therapeutic Target Database | T31204 |
| ChEMBL | CHEMBL1833 |
| IUPHAR | 7 |
| DrugBank | BE0000393 |
| Name | 1-Methyllysergic acid butanolamide |
|---|---|
| Molecular formula | C21H27N3O2 |
| IUPAC name | N-(1-hydroxybutan-2-yl)-4,7-dimethyl-6,6a,8,9-tetrahydroindolo[4,3-fg]quinoline-9-carboxamide |
| Molecular weight | 353.466 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 2.3 |
| Synonyms | AC1L1HJY L001319 (+)-9,10-Didehydro-N-[1-(hydroxymethyl)propyl]-1,6-dimethylergoline-8.beta.-carboxamide Ergoline-8-carboxamide, 9,10-didehydro-N-[(S)-1-(hydroxymethyl)propyl]-1,6-dimethyl-, (8.beta.)- 1-Methyl-D-lysergic acid butanolamide [ Show all ] |
| Inchi Key | KPJZHOPZRAFDTN-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C21H27N3O2/c1-4-15(12-25)22-21(26)14-8-17-16-6-5-7-18-20(16)13(10-23(18)2)9-19(17)24(3)11-14/h5-8,10,14-15,19,25H,4,9,11-12H2,1-3H3,(H,22,26) |
| PubChem CID | 4163 |
| ChEMBL | N/A |
| IUPHAR | N/A |
| BindingDB | N/A |
| DrugBank | N/A |
Structure | 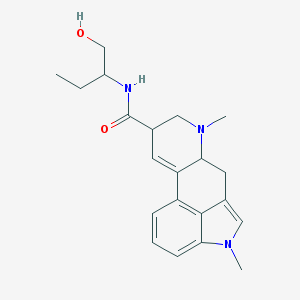 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | 0.1 nM | http://pdsp.med.unc.edu/pdsp.php | PDSP |
| Ki | 0.363078 nM | PMID15322733 | PDSP |
| Ki | 0.64 nM | PMID9459568 | PDSP |
| Ki | 1.51 nM | PMID9459568 | PDSP |
| Ki | 1.59 nM | PMID8632342 | PDSP |
| Ki | 1.65 nM | PMID9225287 | PDSP |
| Ki | 7.94 nM | PMID7582481 | PDSP |
| Ki | 7.94328 nM | PMID10821800 | PDSP |
| Ki | 9.1 nM | PMID11104741 | PDSP |
| Ki | 19.0 nM | http://www.tandfonline.com/doi/pdf/10.1080/07391102.2013.869483 | PDSP |
| Ki | 150.0 nM | http://molpharm.aspetjournals.org/cgi/content/full/63/6/1223 | PDSP |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417