

You can:
| Name | Beta-2 adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRB2 |
| Synonym | beta-2 adrenergic receptor Gpcr7 beta2-adrenoceptor Adrb-2 ADRB2R [ Show all ] |
| Disease | Premature labour Premature ejaculation Obesity Neurogenic bladder dysfunction Hypertension [ Show all ] |
| Length | 413 |
| Amino acid sequence | MGQPGNGSAFLLAPNGSHAPDHDVTQERDEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYANETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRFHVQNLSQVEQDGRTGHGLRRSSKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLCLRRSSLKAYGNGYSSNGNTGEQSGYHVEQEKENKLLCEDLPGTEDFVGHQGTVPSDNIDSQGRNCSTNDSLL |
| UniProt | P07550 |
| Protein Data Bank | 3nya, 3ny9, 3ny8, 3d4s, 2rh1, 3pds, 4gbr, 4lde, 6mxt, 6csy, 5x7d, 5jqh, 5d6l, 5d5b, 5d5a, 4ldo, 4ldl, 4qkx |
| GPCR-HGmod model | P07550 |
| 3D structure model | This structure is from PDB ID 3nya. |
| BioLiP | BL0257082, BL0113951, BL0113950, BL0257084, BL0257085, BL0283869, BL0333729, BL0333730, BL0333731,BL0333732,BL0333733, BL0333734, BL0333735, BL0333736,BL0333737,BL0333738, BL0113952,BL0113953,BL0113954, BL0147310, BL0257081, BL0257080, BL0232997, BL0192129, BL0257083, BL0185746,BL0185747, BL0185745, BL0185743,BL0185744, BL0185742, BL0185740,BL0185741, BL0147311,BL0147312, BL0351701,BL0351703, BL0351702,BL0351704, BL0192128, BL0433200, BL0433199, BL0430930, BL0430929, BL0388810, BL0388809, BL0354449, BL0185748, BL0354450, BL0354451,BL0354452,BL0354453, BL0388807,BL0388808 |
| Therapeutic Target Database | T24555, T52522 |
| ChEMBL | CHEMBL210 |
| IUPHAR | 29 |
| DrugBank | BE0000694 |
| Name | isoproterenol |
|---|---|
| Molecular formula | C11H17NO3 |
| IUPAC name | 4-[1-hydroxy-2-(propan-2-ylamino)ethyl]benzene-1,2-diol |
| Molecular weight | 211.261 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 4 |
| XlogP | -0.6 |
| Synonyms | LS-42869 NCGC00015558-05 NCGC00015558-14 Norisodrine Oprea1_009434 [ Show all ] |
| Inchi Key | JWZZKOKVBUJMES-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C11H17NO3/c1-7(2)12-6-11(15)8-3-4-9(13)10(14)5-8/h3-5,7,11-15H,6H2,1-2H3 |
| PubChem CID | 3779 |
| ChEMBL | CHEMBL434 |
| IUPHAR | 536 |
| BindingDB | 25392 |
| DrugBank | DB01064 |
Structure | 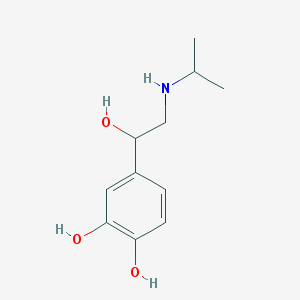 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Activity | 100.0 % | PMID24326276 | ChEMBL |
| EC50 | 0.1585 nM | PMID16640337, PMID11806709 | ChEMBL |
| EC50 | 0.16 nM | PMID16640337, PMID11806709 | BindingDB |
| EC50 | 0.2 nM | PMID24326276 | ChEMBL |
| EC50 | 0.84 nM | PMID19608416 | BindingDB |
| EC50 | 0.84 nM | PMID19608416 | ChEMBL |
| EC50 | 2.0 nM | PMID15603933, PMID18651730, PMID18553954, PMID19366244 | BindingDB,ChEMBL |
| EC50 | 3.0 nM | PMID19232786, PMID19362005, PMID19581100 | BindingDB,ChEMBL |
| EC50 | 39.81 nM | PMID19317397, PMID21696967, PMID21652207, PMID21925889, PMID20462258 | ChEMBL |
| EC50 | 40.0 nM | PMID19317397, PMID21696967, PMID20462258, PMID21925889, PMID21652207 | BindingDB |
| EC50 | 87.0 nM | PMID27132867 | BindingDB,ChEMBL |
| EC50 | 120.0 nM | PMID27132867 | BindingDB,ChEMBL |
| EC50 | 129.0 nM | PMID22182578 | BindingDB |
| EC50 | 240.0 nM | PMID22182578 | BindingDB |
| EC50 | 1000.0 nM | PMID19581100 | BindingDB,ChEMBL |
| Efficacy | 1.0 - | PMID2999409 | ChEMBL |
| Emax | 100.0 % | PMID27132867 | ChEMBL |
| Emax | 116.0 % | PMID16640337, PMID11806709 | ChEMBL |
| IA | 100.0 - | PMID10691685 | ChEMBL |
| IC50 | 246.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Intrinsic activity | 1.0 - | PMID21925889, PMID21696967, PMID21652207, PMID19232786, PMID20462258 | ChEMBL |
| Intrinsic activity | 100.0 - | PMID10377236 | ChEMBL |
| Kd | 50.0 nM | PMID19168263 | BindingDB,ChEMBL |
| Kd | 251.19 nM | PMID23954364 | ChEMBL |
| Ki | 2.44 nM | PMID24326276 | ChEMBL |
| Ki | 46.0 nM | PMID8699, PMID27132867 | BindingDB,ChEMBL |
| Ki | 136.0 nM | PMID18783211 | BindingDB |
| Ki | 169.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Ki | 192.0 nM | PMID24326276 | ChEMBL |
| Ki | 398.107 nM | PMID8982677 | IUPHAR |
| Ki | 458.0 nM | PMID14730417 | BindingDB |
| Ki | 501.0 nM | PMID22182578 | BindingDB |
| Ki | 676.08 nM | PMID10377236 | ChEMBL |
| Ki | 1258.93 nM | PMID16051698 | PDSP |
| Ki | 1258.93 nM | PMID16051698 | BindingDB |
| Ki | 1513.56 nM | PMID7915318 | PDSP,BindingDB |
| Ki | 2800.0 nM | PMID8699 | BindingDB |
| Km | 50.0 nM | PMID2999409 | ChEMBL |
| pK | 9.46 - | PMID10691685 | ChEMBL |
| pKact | 8.4 - | PMID10377236 | ChEMBL |
| Potency | 1299.5 nM | PubChem BioAssay data set | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417