

You can:
| Name | Muscarinic acetylcholine receptor M2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM2 |
| Synonym | cholinergic receptor AChR M2 M2 muscarinic acetylcholine receptor M2 receptor Chrm-2 [ Show all ] |
| Disease | Urinary incontinence Heart failure Nausea; Addiction Parkinson's disease Peptic ulcer [ Show all ] |
| Length | 466 |
| Amino acid sequence | MNNSTNSSNNSLALTSPYKTFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRIKKDKKEPVANQDPVSPSLVQGRIVKPNNNNMPSSDDGLEHNKIQNGKAPRDPVTENCVQGEEKESSNDSTSVSAVASNMRDDEITQDENTVSTSLGHSKDENSKQTCIRIGTKTPKSDSCTPTNTTVEVVGSSGQNGDEKQNIVARKIVKMTKQPAKKKPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLMCHYKNIGATR |
| UniProt | P08172 |
| Protein Data Bank | 5zkc, 4mqs, 4mqt, 5yc8, 5zk3, 5zkb, 5zk8 |
| GPCR-HGmod model | P08172 |
| 3D structure model | This structure is from PDB ID 5zkc. |
| BioLiP | BL0433341, BL0433216, BL0263147, BL0263146, BL0263145, BL0433339, BL0433340, BL0433338 |
| Therapeutic Target Database | T46185 |
| ChEMBL | CHEMBL211 |
| IUPHAR | 14 |
| DrugBank | BE0000560 |
| Name | arecoline |
|---|---|
| Molecular formula | C8H13NO2 |
| IUPAC name | methyl 1-methyl-3,6-dihydro-2H-pyridine-5-carboxylate |
| Molecular weight | 155.197 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 0 |
| XlogP | 0.3 |
| Synonyms | Arecaline NCGC00015075-05 BBL036229 Nicotinic acid,2,5,6-tetrahydro-1-methyl-, methyl ester BSPBio_002974 [ Show all ] |
| Inchi Key | HJJPJSXJAXAIPN-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C8H13NO2/c1-9-5-3-4-7(6-9)8(10)11-2/h4H,3,5-6H2,1-2H3 |
| PubChem CID | 2230 |
| ChEMBL | CHEMBL7303 |
| IUPHAR | 296 |
| BindingDB | 46858 |
| DrugBank | DB04365 |
Structure | 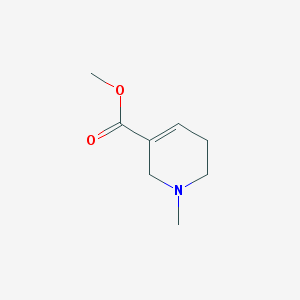 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 25.0 nM | PMID7783150 | BindingDB,ChEMBL |
| EC50 | 83.0 nM | PMID7990109 | BindingDB,ChEMBL |
| EC50 | 10000.0 nM | PMID9873644 | BindingDB,ChEMBL |
| IC50 | 1800.0 nM | PMID9435896 | BindingDB,ChEMBL |
| IC50 | 4500.0 nM | PMID9438027 | BindingDB,ChEMBL |
| Ki | 2.4 nM | PMID12747793 | PDSP |
| Ki | 40.0 nM | PMID9622546, PMID10891110 | BindingDB,ChEMBL |
| Ki | 2400.0 nM | PMID12747793 | BindingDB,ChEMBL |
| Ki | 2454.71 nM | PMID18182302 | ChEMBL |
| Ki | 6309.57 nM | PMID9224827 | IUPHAR |
| Max cAMP | 119.0 % | PMID9873644 | ChEMBL |
| Ratio | 0.8 - | Bioorg. Med. Chem. Lett., (1991) 1:3:147 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417