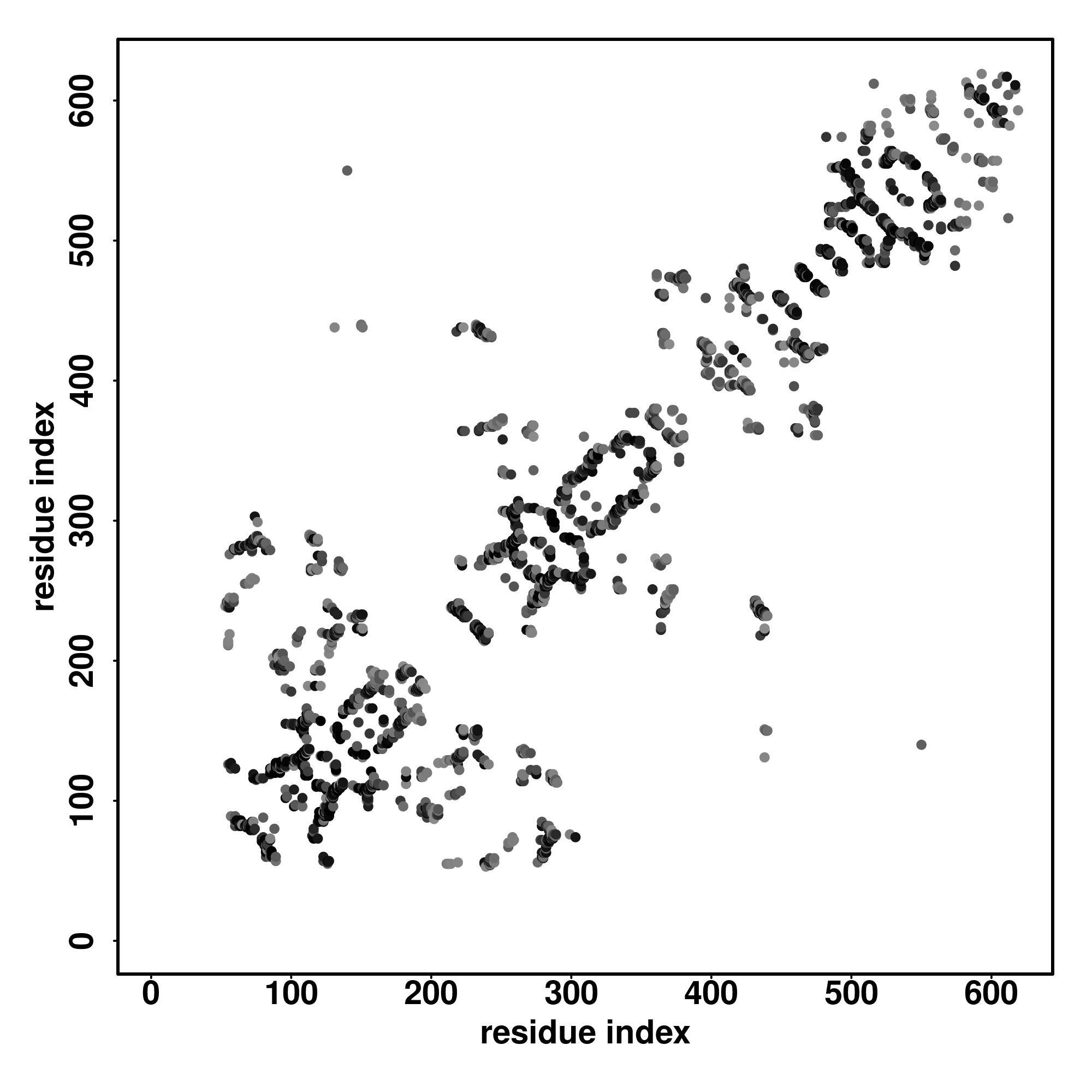
| Rank | PDB
hit | ID1 | ID2 | Cov | Norm.
Zscore | Download
alignment | | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 480
| | | | | | | | | | | | | | | | | | | | | | | | |
|---|
| SS
Seq | CCCHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHCCCCSSSSSCCSSSSHHHHHHHHHHHHHHHHHHHCCCCCCSSSSSCCCCHHHHHHHHHHHHHCCSSSSCCCCCCHHHHHHHHHHCCCSSSSSCHHHHHHHHHHHHHCCCCSSSSSCCCCCCCCCCCHHHHHHCCCCCCCCCCCCCCCCCCCSSSSSCCCCCCCCCCSSSSCHHHHHHHHHHHHHHCCCCCCSSSSSCCCHHHHHHHHHHHHHHHHCCSSSSCCCCCHHHHHHHHHHHCCCSSSCHHHHHHHHHHCCCCCCCCCCCSSSSSSCCCCHHHHHHHHHHHCCCCSSSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSSSSCCCCCCSSSCCCCCSSSCCCCCCSSSSSSSCCCCCCCCCCCCHHHHHHHHHHHCCCCCCSSSSCCCSSSSCCCCCSSSSCCCC
MLLSWLTVLGAGMVVLHFLQKLLFPYFWDDFWFVLKVVLIIIRLKKYEKRGELVTVLDKFLSHAKRQPRKPFIIYEGDIYTYQDVDKRSSRVAHVFLNHSSLKKGDTVALLMSNEPDFVHVWFGLAKLGCVVAFLNTNIRSNSLLNCIRACGPRALVVGADLLGTVEEILPSLSENISVWGMKDSVPQGVISLKEKLSTSPDEPVPRSHHVVSLLKSTCLYIFTSGTTGLPKAAVISQLQVLRGSAVLWAFGCTAHDIVYITLPLYHSSAAILGISGCVELGATCVLKKKFSASQFWSDCKKYDVTVFQYIGELCRYLCKQSKREGEKDHKVRLAIGNGIRSDVWREFLDRFGNIKVCELYAATESSISFMNYTGRIGAIGRTNLFYKLLSTFDLIKYDFQKDEPMRNEQGWCIHVKKGEPGLLISRVNAKNPFFGYAGPYKHTKDKLLCDVFKKGDVYLNTGDLIVQDQDNFLYFWDRTG |
|---|
| 1 | 7kdsA | 0.19 | 0.17 | 5.46 | 1.33 | DEthreader | | -------------------FE---KGIHDVNQYKQMYQSFFGPLAKFVFLGGELNASYNCDRHAFANPDKPALICESHILTYGDLLREVSKVAGVLQS-WGIKKGDTVAVYLPMNAQAIIAMLAIARLGAAHSVIFAGFSAGSIKDRVNDASCKALITCDEIKKLCDEALVDCPTVEKVLVYKNIHLTRDYYWDVETAKFPG-YLPP--VS-VNSEDPLFLLYTSGSTGTPKGVVHSTAYLLGAALSTKYFDIHPEDILFTAGDVGWITGHTYALYGPLLLGVPTIIFEGTPYGRFWQIVEKHKATHFYVAPTALRLLRGEQEIAKYDLSSLRTLGSVPISPDIWEWYNEFGKQCHISDTYWQTESGSHLAPLAVPNKPGSASY--P-FFG-IDAALIDP--VTGV-------EIEGNDAEGVLAIKDHWPSMARTVYKNHTKYMDTYMNPYP---G-YYFTGDGAARDHDGYYWIRGRVD |
| 2 | 1mdfA1 | 0.17 | 0.15 | 4.79 | 2.18 | SPARKS-K | | -------------------------------MLKGFTPWPDELAETYRKNGAGETFGDLLRDRAAKYGDRIAITCGNTHWSYRELDTRADRLAAGFQKLG-IQQKDRVVVQLPNIKEFFEVIFALFRLGALPVFALPSHRSSEITYFCEFAEAAAYIIPDAYRSLARQVQSKLPTLKNIIVAGE--AEEFLPLEDLHTEPVKLPEVK-------SSDVAFLQLSGGSTGLSKLIPRTHDDYIYSLKRSVEVWLDHSTVYLAALPMAHNYPLSSGVLGVLYAGGRVVLSPSPSPDDAFPLIEREKVTITALVPPLAMVWMDAASSRRDDLSSLVLQVGAKFSAEAARRVKAVFG-CTLQQVFGMAEGLVNYTRLDDPVNTQGKPMSPYDESR--------------VWDDHDR--DVKPGETGHLLTRG--PYTIRGYYKAEEHNAASFT------EDGFYRTGDIVRLTRDGYIVVEGRAK |
| 3 | 7kq6A | 0.19 | 0.17 | 5.65 | 0.76 | MapAlign | | -------LSGMNEYNQLYQQSINDPDTFWARMARDLTHIGTLEGDNAWFVGGRLNASFCVDRHAMRDPNKVAIIYEGRSITYAELLKEVSRLAWVMKS-QGVRKGDTVAIYLPMIPEAIFALLACARIGAIHSVVFAGFSSDSLRDRTLDARSKFIITTDITKKIVDEALKQC-PDVNCLVFKRTVPGRDLWWHEEVDKY-PNYL---PAESMDSEDPLFLLYTSGSTGKPKGVMHTTAYLVGAAATGKVFDIHPADRFFCGGDVGWITGHTYVVYAPLLLGCTTVVFESPAFSRYWDVIEKHKVTQFYVAPTALRLLKRAGDHHINHEMDLRILGSVPIAAEVWKWYHEVGRQAHIVDTYWQTETGSHITPLGTKPGSASLP------FFGIDPVILDPVTGAEI---------PGNDVEGILAFRKPWPSMARTVWGDHKRYMDTYLNVY----KGFYFTGDGAGRDHEGYYWIRGRVD |
| 4 | 6m2oA | 0.16 | 0.14 | 4.49 | 0.38 | CEthreader | | ----------------------------------------------VTPPPEKFNFAEHLLQTNRVRPDKTAFVDDISSLSFAQLEAQTRQLAAALRA-IGVKREERVLLLMLDGTDWPVAFLGAIYAGIVPVAVNTLLTADDYAYMLEHSRAQAVLVSGALHPVLKAALTKSDHEVQRVIVSRPAAPLEPGEVDFAEFVGAHAPLEK-PAATQADDPAFWLYSSGSTGRPKGVVHTHANPYWTSELYGTLHLREDDVCFSAAKLFFAYGLGNALTFPMTVGATTLLMGRPTPDAVFKRWLGVKPTVFYGAPTGYAGMLAAPNLPSRDQVALRLASSEALPAEIGQRFQRHFG-LDIVDGIGSTEMLAAFLSNLVRYGTTGWPV-------PGYQIELRGDGGG----------PVADGEPGDLYIHG--PSSATMYWGNRAKSRDTF-------QGGWTKSGDKYVRNDDGSYTYAGRTD |
| 5 | 3etcB1 | 0.20 | 0.18 | 5.78 | 1.72 | MUSTER | | ----------------SLLSQFVSKTDFESYEDFQENF-------KILVPENFNFAYDVVDVYARDSPEKLAMIWCDKIFTFKDLKYYSDKAANFFVKH--IGKGDYVMLTLKSRYDFWYCMLGLHKLGAIAVPATHMLKTRDIVYRIEKAGLKMIVCIAEVPEQVDEAHAECGDILKKAKVGGDVLEGWIDFRKELEESSPIFERPTGEVSTKNEDICLVYFSSGTAGFPKMVEHDNTYPLGHILTAKYQNVEDDGLHYTVADSGWGKCVWGKLYGQWIAGCAVFVYDRFEAKNMLEKASKYGVTTFCAPPTIYRFLIKE-DLSHYNFSTLKYAVVEPLNPEVFNRFLEFTGIK-LMEGFGQTETVVTIATFPWMEP---KPGSIGKPTPGYKIELMDRDGR-----------LCEVGEEGEIVINGKPVGLFVHYGKDPERTEETW-------HDGYYHTGDMAWMDEDGYLWFVGRAD |
| 6 | 3tsyA | 0.20 | 0.18 | 5.63 | 1.37 | HHsearch | | -------------------------------------DVI-FRSKLPIYIPNHLSLHDYIFQNISEFATKPCLINGGHVYTYSDVHVISRQIAANFH-KLGVNQNDVVMLLLPNCPEFVLSFLAASFRGATATAANPFFTPAEIAKQAKASNTKLIITEARYVDKIKPLQNDDG--VVIVCIDDNEPEGCLRFTELTQSTTEA-SEVIDSVEISPDDVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVNLYFHSDDVILCVLPMFHIYALNSIMLCGLRVGAAILIMPKFEINLLLELIQRCKVTVAPMVPPIVLAIAKSSETEKYDLSSIRVVKAAPLGKELEDAVNAKFPNAKLGQGYGMTEAGPVLMKEPVKSGACGTVV------RNAEMKIVDPDTG----------DSLSRNQPGEICIR--GHQIMKGYLNNPAATAETI------DKDGWLHTGDIGLIDDDDELFIVDRLK |
| 7 | 1ba3A1 | 0.17 | 0.14 | 4.60 | 3.59 | FFAS-3D | | ---------------------------------------------------EDGTAGEQLHKAMKRVPGTIAFTDIEVNITYAEYFEMSVRLAEAMKR-YGLNTNHRIVVCSENSLQFFMPVLGALFIGVAVAPANDIYNERELLNSMNISQPTVVFVSKKGLQKILNVQKKLPIIQKIIIMDSKTDYQGFQSMYTFSHLPPGFNEYDFVPESFDRDKTIALIMNSSTGLPKGVALPHRTACVRFSHARDGNQIIPDTAILSVVPFHHGFGMFTTLGYLICGFRVVLMYRFEEELFLRSLQDYKIQSALLVPTLFSFFAKSTLIDKYDLSNLHEIAGAPLSKEVGEAVAKRFHLPGIRQGYGLTETTSAILITPEGDDKPGAVGK---VVPFFEAKVVDLDTGK----------TLGVNQRGELCVR--GPMIMSGYVNNPEATNALI------DKDGWLHSGDIAYWDEDEHFFIV---- |
| 8 | 6n8eA | 0.12 | 0.11 | 3.84 | 0.95 | EigenThreader | | GARAEALVEAVDTGLAGMRRHQR-YRAEDIRRDCELARWIERFVAFTTAFVADPSCVEVFERRVAERPHASAVTLDHTTWDYAELDARANRLARHFAASTPARGNLRVALLLPRTLDAIVAILATLKFGAAYVPIDPDAPAERIRAIIDDCDAALVVTTVDLASRI------DASGRRLVVLDA-------PDTRAAVAAASAAPPSRDGEGPRADDLAYIIFT---SKPKGVKITHRNVVRLFEATDAWFHYRDDDVWTMCHAYVFD-ASVWEMWGALLHGGRLVVVPTRAPDALLELVVREGVTVFGQIPSAFYRFMEAQADHQALRLRYQCFGGEALDPSRLKPWFDWHRRLLNMYGITETTINATYRFIDRGSLIGEVYA-------DLGIVVLDDALRPV-----------PAGAYGEMYVTGAGLA---QGNRPDLDAVRFVANPYGPAGTRMYRSGDVARLHPDGVLEYVGADQ |
| 9 | 3etcA | 0.21 | 0.19 | 5.92 | 2.59 | CNFpred | | ------------------------------------YEDFQENFKILVP-ENFNFAYDVVDVYARDSPEKLAMIWCEKIFTFKDLKYYSDKAANFFVKH-GIGKGDYVMLTLKSRYDFWYCMLGLHKLGAIAVPATHMLKTRDIVYRIEKAGLKMIVCIADVPEQVDEAHAECGIPLKKAKVGGDVLEGWIDFRKELEESSPIFERPTGEVSTKNEDICLVYFSSGTAGFPKMVEHDNTYPLGHILTAKYWQVEDDGLHYTVADSGWGKCVWGKLYGQWIAGCAVFVYDYFEAKNMLEKASKYGVTTFCAPPTIYRFLIKEDL--NFSTLKYAVVAGEPLNPEVFNRFLEFTG-IKLMEGFGQTETVVTIATFPW---MEPKPGSIGKPTPGYKIELMDRD-----------GRLCEVGEEGEIVINTKPVGLFVHYGKDPERTEETWH-------DGYYHTGDMAWMDEDGYLWFVGRAD |
| 10 | 7kcpA | 0.19 | 0.17 | 5.46 | 1.33 | DEthreader | | -------------------YE---EPLSGMNEYNQLYQSFWARMARFTFVGGRLNASFNCDRHAMRDPNKVAIIYEGRSITYAELLKEVSRLAWVMK-SQGVRRGDTVAIYLPMIPEAIFALLACARIGAIHSVVFAGFSSDSLRDRTLDARSKFIITTDETKKIVDEALKQCPDVTSCLVFKGVWTKRDLWWHEEVEKYPN-YLPA--ES-MDSEDPLFLLYTSGSTGKPKGVMHTTAYLVGAAATGKYFDIHPADRFFCGGDVGWITGHTYVVYAPLLLGCTTVVFESTPFSRYWDVIEKHKVTQFYVAPTALRLLKAGDHHINHEMKDLRILGSVPIAAEVWKWYHEVGKQAHIVDTYWQTETGSHITPLGTPTKPGSASL--P-FFG-IDPVILDP--VTGA-------EIPGNDVEGILAFRKPWPSMARTVWGDHKRYMDTYLNVYK---G-FYFTGDGAGRDHEGYYWIRGRVD |
| (a) | ID1 is the number of template residues identical to query divided by number of aligned residues. |
| (b) | ID2 is the number of template residues identical to query divided by query sequence length. |
| (c) | Cov is equal the number of aligned template residues divided by query sequence length. |
| (d) | Norm. Zscore is the normalized Z-score of the threading alignments. A Normalized Z-score >1 means a good alignment and is highlighted in bold. |
| (e) | Download alignment lists the threading program used to identify the template, and provide the 3D structure of aligned regions of threading templates (threading[1-10].pdb.gz). |
| (f) | Template residues identical to query sequence are highlighted in color. |
|