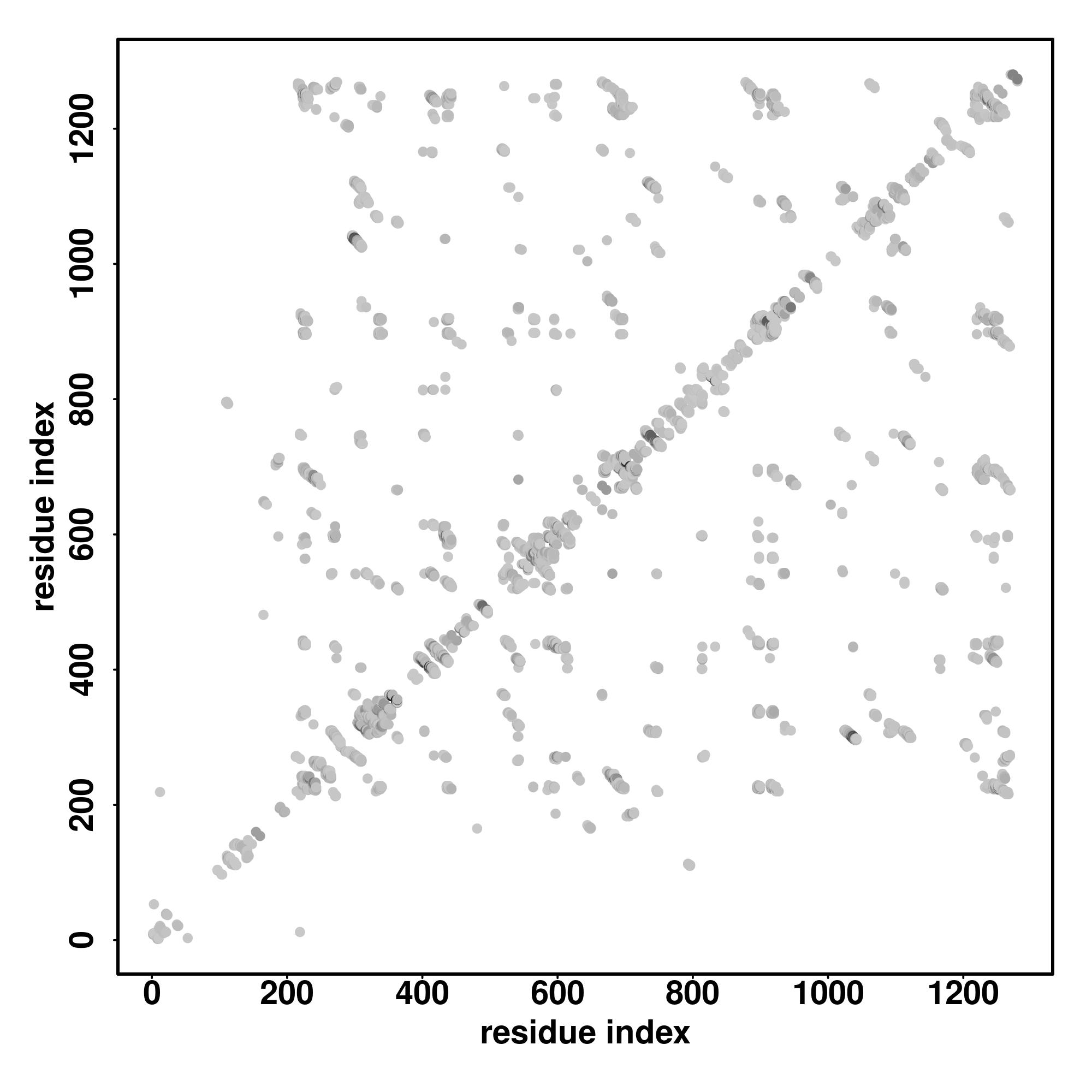
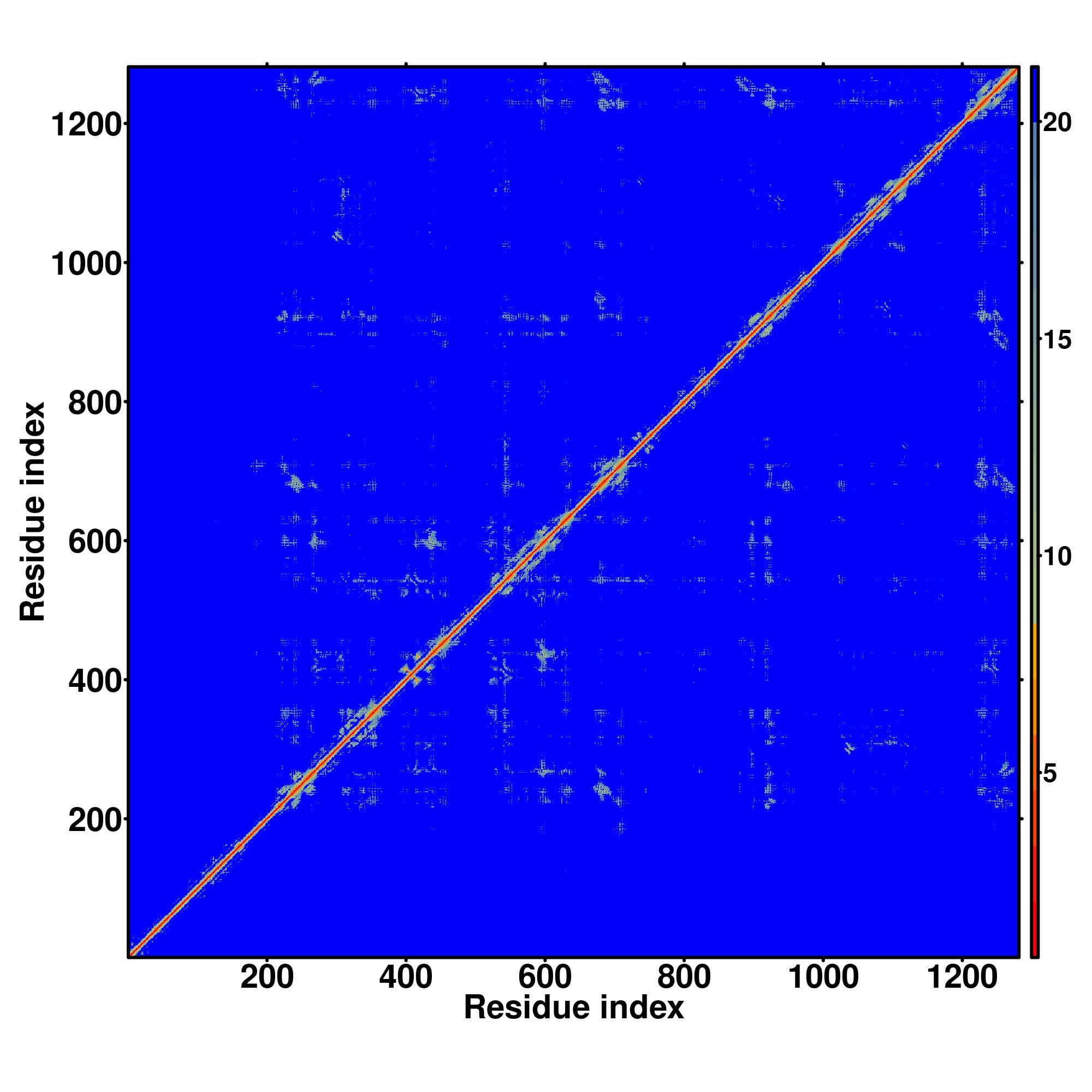
| Rank | PDB
hit | ID1 | ID2 | Cov | Norm.
Zscore | Download
alignment | | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 480 500 520 540 560 580 600 620 640 660 680 700 720 740 760 780 800 820 840 860 880 900 920 940 960 980 1000 1020 1040 1060 1080 1100
| | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | |
|---|
| SS
Seq | CCCCCCCCCCCCCCCCCCCSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCCHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHCCCCCCCCCCCHHHHCCHHHHHHHHHHCCCCCCCCCCCCCSSSCCCCCCSSSCCCCCCCCCCCCCSSCCCCCCCCCCCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCHHHCCCCCCSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSCCCCCCCHHHHCCCCCCCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCHHHHHCCCCCCCCCCCCCCCHHHHHHCCCCCCCCCCCCCCSSCCCCCCCCCCHHHHHHHHHHHCCCCCCHHHHHHCCCCCCCCCCCCCCCCCCSCCCCCCSCCCHHHHHHHHHCCCCCSSCCCCCCCCCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSCCCCCCCCCCCCCCCCCCCSCCCCHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCCC
MVRKKNPPLRNVASEGEGQILEPIGTESKVSGKNKEFSADQMSENTDQSDAAELNHKEEHSLHVQDPSSSSKKDLKSAVLSEKAGFNYESPSKGGNFPSFPHDEVTDRNMLAFSSPAAGGVCEPLKSPQRAEADDPQDMACTPSGDSLETKEDQKMSPKATEETGQAQSGQANCQGLSPVSVASKNPQVPSDGGVRLNKSKTDLLVNDNPDPAPLSPELQDFKCNICGYGYYGNDPTDLIKHFRKYHLGLHNRTRQDAELDSKILALHNMVQFSHSKDFQKVNRSVFSGVLQDINSSRPVLLNGTYDVQVTSGGTFIGIGRKTPDCQGNTKYFRCKFCNFTYMGNSSTELEQHFLQTHPNKIKASLPSSEVAKPSEKNSNKSIPALQSSDSGDLGKWQDKITVKAGDDTPVGYSVPIKPLDSSRQNGTEATSYYWCKFCSFSCESSSSLKLLEHYGKQHGAVQSGGLNPELNDKLSRGSVINQNDLAKSSEGETMTKTDKSSSGAKKKDFSSKGAEDNMVTSYNCQFCDFRYSKSHGPDVIVVGPLLRHYQQLHNIHKCTIKHCPFCPRGLCSPEKHLGEITYPFACRKSNCSHCALLLLHLSPGAAGSSRVKHQCHQCSFTTPDVDVLLFHYESVHESQASDVKQEANHLQGSDGQQSVKESKEHSCTKCDFITQVEEEISRHYRRAHSCYKCRQCSFTAADTQSLLEHFNTVHCQEQDITTANGEEDGHAISTIKEEPKIDFRVYNLLTPDSKMGEPVSESVVKREKLEEKDGLKEKVWTESSSDDLRNVTWRGADILRGSPSYTQASLGLLTPVSGTQEQTKTLRDSPNVEAAHLARPIYGLAVETKGFLQGAPAGGEKSGALPQQYPASGENKSKDESQSLLRRRRGSGVFCANCLTTKTSLWRKNANGGYVCNACGLYQKLHSTPRPLNIIKQNNGEQIIRRRTRKRLNPEALQAEQLNKQQRGSNEEQVNGSPLERRSEDHLTESHQREIPLPSLSKYEAQGSLTKSHSAQQPVLVSQTLDIHKRMQPLHIQIKSPQESTGDPGNSSSVSEGKGSSERGSPIEKYMRPAKHPNYSPPGSPIEKYQYPLFGLPFV |
|---|
| 1 | 6pbcA | 0.06 | 0.05 | 2.05 | 1.25 | EigenThreader | | KTSRDFDRYQEDPAFRPDQSHEFRLKTSLQATSEDEVNMWIKGLTWLMEDTLQ------------AATPLQIERWLRKQFYSVDRAKDLKNMLSQVNYRVPDITYGQFAQLYRSLMYSAQKTMD--------------LPFCQVSEFQQFLLEYQGELWAVDRLQVQEFMLSFLRDIEEPYFFLDELVTFLFSKENSVWNSQLDAVCPETMNNPLS----HNTYLTGDQFSSESSLEAYARCLRM----------------------------------------------------GCR---CIELDCWDGPDGMPVIYHGHTLTTKPVILSIEDHCSIAQQRNMAQHFRKVLGDTLLT----KPVDIAADGLPSPNQLKRKKKLAEGSNDISNSIKNGILYLEDPVNHEWYPHYFVS---------SKIYYSEETLHS--------------------SEKWFHGKLGAGRDGRHIAERLLTEYCIETGAPDGSFLV------RESETFVGDNGKVQHCRIHSRQPKFFLTDNLVFDS---------LYDLITHYQQVPLRFEMRLSEPVPQTNAHESKEWYHASL---TRAQAEHMLMRVPRDG---AFLVRKRNEP---------------------------------------NQEGQTVMLGNFDSLVDLISYYEKH---------PLYRKMKLRNEEALEKIAVKALFDYKAQREDELTFTKSAIIQNVEKQDGGWWRGDYGGNPAILEPEREH---------------------LDENSPLGDLLRGVLDVPACEGKNNRLFVFSISMPSV--------AQWSLDVAADSQEELQDWVKKIREVAQTKIALELSELVVYCPFDEEKIGTE------------------------------RACYRDMSSFPETKAEKYVNKAKGKKFLQYNRLQLSR--IYPKGQR--------LDSSNYDPLPMWICGLNFQTPDKPMQMNQALFMAGGHCGYVLQPSTMRDEAFDPFD------KSSLR--GLEPGARHLPKNGRGIVCEYDSEFVVDNGLNP---------VWPAKPNPEFMFSDPVKGLKTNNYSED |
| 2 | 6zywY | 0.08 | 0.07 | 2.60 | 1.23 | SPARKS-K | | VMKPTKKVGILLGQKDKGKI-----NSIEKWPLIQSYGLEELGVGFEVVDLTLRLNAVYKNY---DKFFVSKLIYVVAKRLTGHFNSAAGQLGDMKMHKRNLA-TESQLTEIFRDTYEIEEISKWVQIRGVNAALPKPRVLKNTSADCSKEPSVAPLKDLKYSETFHSFHATFETFDLRTCLRAARTYFLAKGVKEER--NLITLNDDEGVPQGYE------------------LNIDENQQYKDQDFLANLYLSIIIGFNEVMQLITKDYKNMTEEFIQDYIFQKVSKVYAEITLDKI-----QIILKAYNSF-----GEEVKI-------------------------------------DFKDTISFKLTPYFFMVRIEQKNIKSQILNNTVLGSLVFAESFILQ------------EGCYLLLTKEIP----YFDLWNCQN----DYSEKIEK------------------MKKRILWEPLGKQISDELPKNRIFVQTGRKSNYGFDIPIMQASYYMRIETRLGWFILFFKEMKEIQITQK------MNHTWLIFKVD------SNITFNSISKDTIALEFTALEQSFFKIKNYFEENQIKYEYQVDIPAIFQESQIAKQFFISYIESKQLMILNQMKDLKNLYEQMQISQAITPVEN-----HIGVILVNG-----------SYCS-----------GKRKFAENLIRF-----------GSDNNLRLHLYKF------------DLNEMSELTEKSYLSGLLKFASEKKIQNTDVIVASVPHFINTKILIDYFSKSEKISNAFYIRTIATNINNIYSNFNKNPVNNVFTYGVEGYSQFLLLDTNKTLSGVLPGAKIY-KIMNNILNPTSITFISEQNNLNRLKYSVQYD-----------LLTSNGP--------SSVVFIPFKLPIL-------REKIRDLIYKKILQNGQAIKIDYVKGILRYDSKLKEGLEEITITPNYFIERTVKGVDAKEFTEELNGVSFKNVKYT-----GITNSIINDMGFVFAGKNLNKELVKPLNKQKLRQRKDLTEEEIVDIQFRNREGLENGEFYDGQFWLILHHPKKDEFIE |
| 3 | 6rc9A | 0.07 | 0.05 | 2.07 | 2.32 | MapAlign | | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------NPRLTPWTYRNTSFSSPLTGENPGAWALVRDNSAKGITSQQTTYDPTRTEAALTAS-------------------------------TTFALRRYDLAGRALYDLDFSKLNPVPVEVAQDPSNPRFAVLLVPRSQQRTESGSTTGAMFGLKVKNAEADTAKSNEKLQGAPIKRSEESGQSVQLKADDFGTALSPTPWASILILYDAPYARNRTAIDRVDHLDPPKWNHHGLWDWKARDVQTTGFFNPRRHPEWFDGG--QTVADNEKTGFDVDNSENTKQGFQKEADSDKSAPIALPF--------EAYFANIGNLTWFGQALLVFGGNGTKSAHTAPLSIGVFRVRYNATGTSATVTGWPYDGLKDLPFNNNRWFEYVPRMAVAGAKVGRELVLAGTITMLLYDELESNLNLVAQGQGLLYGWANRPDLPIGAWSSSSSSSHNAPYYFHNNPDWQDRPQNVVDAWQVYNWSNKLTDQPLSADFVAANEYERFNLVGSVLDQVLDYVPWIGNGYRYGNNHRG-VDDITAPRSFLPTFSNIGV------GLKANVQATLNLQLWTGAGWASSGQSDEDSGDSLTTQDGNAIDQQEATSFTNKNNAQRAQLFLRGLLGSIPVLVNRSGSDSNKFQATDQKWSYTDLHSDQTKLNLPAYGEVNGLLNPALVETYFLVVSGTTVSFQRPWAAFRGSWVNRLGRVESVWDDDLKNLLDPNQVSSVLSLSPVEKVSGWLVNDVVGVGRLSESNAAKMNDVDGIVRTPLAELLDGEGQTADTGPQSVKFKSPDQIDFNRLFTHPVTDLFDPVTMLVYDQYIPLFIDIPASVVRLKVLSFDTNEQSLGLRLE-FFKPDQDGDFLPLLTASSQGP------------------- |
| 4 | 6djyB | 0.11 | 0.10 | 3.67 | 1.19 | MUSTER | | SDTTGNPRITNATNNDETHATGPIEDLNSTSHGREPE----IESFADRAELAMMIQGMTVGALTVQPMRSIRSTFANLANVLIFHDVFTTEDKPSAFIEYHSDEMI-VNMPKQTYNPIDNLAKILYLPSLEKFKYGTGIVQLNYSPHISKLYQNTNNIINTITDGITYANRTEFFIRVMVLMMMDRKILTMEFYDVDTSAINTAILPTIPTTTGVSPLL--RIDTRTEPIWYNDAIKTLIT---TIQYGKIKTVLDANAVKRYSVVGYPIDQYRAYLYNHNLLE--YLGKKVKREDIMSLILSYEFDLITISDLEYQNIPKWFSDNDLS---------RFIFSICMFPDIVRQFHALNIDYFSQTVKSENAIVKMLNSNQNMEPTIINWFLFRICAIDKTVIDDY--SLEMTPIIMRPKLYDFDMKRGEPVSLLY-ELILFSIMFPNVTQHMLGQIQARILYISMYAFRQEYLKFIT----KFGFYYKIVNGRKEYIQVTNQNERMTENNDVLTGNL-----YPSLFTDDPTLSAIAPTLAKIARLMK-------------------PTTSLTPDDRAIAAKFPRFKD----SAHLNPYSSLNIGGRTQHSVTYTR--------MYDAIEEMFNLILRAFASSFAQRPRAGVTQLKSLL-----------TQLADPLCLALDGHVYHLYNVM--------ANMMQNFIPNTDGQFHSFRACSKDGGNRVVQNGDELNESLLIDTAIWGLL---GNTDSSYGNAIGATGTANVPTKVQPVIPTPDNFITPTIHLKTSIDSVEGSRQTTIPGYEDELNKLRTGISQPKVTERQYRRARESIKNMLGSGDYNVAPLHFLLHTEHRSTKLSKPRRVLDNVVQPYVANLDPAEFENTPQLIENSNMTRLQIALKMLTGDMDDIVKGLILKRAAKFDVYYYVFLIDGVKILAEDIKNQIDITGIWPEINNGFNTITADVRQFMNTTKAETLLISNKSIVHEIMDNALQPKMSSDTLALSEAVYRTIWNSSIITQRISARGL-MNLEDARPPEAKISHQSELDMGKIDETSGEPI-----YTSGLQKMQSSKVSMANVVLSAGSDVI |
| 5 | 4gatA | 0.40 | 0.02 | 0.70 | 1.94 | HHsearch | | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------MKNGEQGPTTCTNCFTQTTPLWRRNPEGQPLCNACGLFLKLHGVVRPLSLKT-----DVIKKRNRNSANS------------------------------------------------------------------------------------------------------------------------------------------------- |
| 6 | 6ajfA | 0.07 | 0.05 | 2.25 | 1.03 | EigenThreader | | ----------------FQSNAMFAWWGRTVYQFRYIVIGVMVALCLGGGVYGISLGNHVTQSGFYDEGSQSVAASLIGDEVYGRDRTSHVPPDDKKVTDKAWQKKVTEELDQVVKDHEDQIVGWLKAPDTTDPTVSAMKTQDLRHTFISIPLQGDDDDEILKNYQVVEPELQQVNGGDLNPLASELTGTIGEDQKRAEVA-----------------------------------AIPLVAVVLFFVFGT----VIAAALPAIIGGLAIAGALGIMRLVAEFTPVHFFAQPVVTLI-------------------------GLGIAIDYGLFIVSRFREEIAEGYDTEAAVRRTVMTSGRTVVFSAVIIVASSVPLLLFPQ-------------------------------------GFLKSITYAIIASV-----MLAAILSITVLAAALAILGPRVDALGVTTLL--------KGFWGRLVNVVMK------RPIAFAAPILVVMVLLIIPLGQLSLGG--------ISEKYLPPDN---AVRQSQEQFDKLF--------------PGFRTEPKREDGEP--ITDAQIADMRAKALTVSGFTDPDNDPWKERPANDSGLENR----------------NDAAKKIDELRALQPPHGVGG-TPALEQDSIHSLFDKLPLMALILIVTTTVLMFLAFGSVVLPIKAALMSALTLGSTMGILTWMFVDGHGSGLMN--YTPQPLMAPMIGLIIAVIWGLSTDYEVFLVSRMVEARERG----MSTAEAIRIGTATTGRLITGAALILAVVAGAFVFSDLVMMKYLAFGLLIALLLDATIIRMFLVPAVMKLLGDDCWWA-----PRWMKRVQEKEFNIFEMLRIDE---GLTEGYYTIGIGHLLTK---SPSLNAAKSELDKAIGRNTNGVITAEKLFNQDVDAAVRGILRNAKLKPVYDSLDAVRRAALINMVFQMGETGVAGFTNSLRMLQQKRWDEAAVNLAKSRWYNQTPNRAKRVITTFRTG-------------------------------------TWDAYEFHLGGIKAFHHHHHHHH------------ |
| 7 | 7abiA | 0.15 | 0.15 | 4.89 | 1.15 | SPARKS-K | | RCWKANIPWKVPGLPTPIENMAKADWWTNTAHYNRERIRRGATVDKTVCTRLYLKAEQERQHNYLKDGPYITAEEAVAVYTTTVHWLESRRFSPIPFPPYKHDTKLLILALERLKEAYSVKSRL-NQSQREELGLIEQAYDNPHEALSRIKRTQRAFKEVGIEFMDLYSHLVPVYDVEPLEKADKRRLFPPWIKPA-DTEPPPLLVNNLQDVWETSEG----ECNVMLESRFEKEKIDLTLLNRLLRLIV------DHNIADYMTAKNN--VVINYKDMNTNSYGIIRGLLLVLGLHRASEMAGP---------------PQMPNDFLSFQFCRYIHIFFRFTADEARDLIQRYLTEHPD--PNNENIVGYNNKKCWPRDARMRLMRLPRSVTTVQWENSFVSVYSKDNPNGFECRILPKCRTSYEEFTHKDGVWRTAQCFLRVDDESMQRFHNRVRQILMASGSTTTQELLDLLVKCENKIQTRTPKELGGLGMLSMGHVL--IPQSDLRWSKQTDVGITHFRDQLNLYRYIQPWESEFIDSQRVWAEYALKRQEAIAQNRRLTSWDRGIPRINTLFQKDRHTLAYDKGWRVRTDFKQYQVLKQNPFW--WTHQRHDGKLWNLNVEGILEHFKGTYFPTWEGLSPTINRANVYVGQVQLDLTGIFMHGKIPTLKISLIQIFRLWQKIHESIVMDLCQVFDQELDAL--EIETVQ-KETIHPRSYKMNSSCADILLFASYKWNVKYWIDIDYDSHDIERYARAKFLDYTTDNMSIYPSPTGVLIAIDLAYNLHSAYGNWFPGSKPLIQQAMIWFVDNVYRVTIHKTFEGNLTINGAIFIFNPRTGQLFLKIIHTSVWAGQKPVEEQPKQIIVTRKGMLDPLEVHLLDVIKGSQLPFQACLKVKFGDLILKATEPQM-VLFNLYDDWLKTISSYTAFSRLIALHVNNDRAKVILKPDIWPTADYGKKNNVNVRDIILGSNYETQTFSSKTEAISAANLHLRTNHIYVSSDDIKSDLRAQGYLYGQVKEIRCIMVPQHQTVHLPGQLPQHEYLKEMEPLGTQPNEQLSPQDVTTHAADNPSWDGEKTIIITCSFTPGSCTLT |
| 8 | 2pn5A1 | 0.07 | 0.04 | 1.62 | 1.89 | MapAlign | | ----------------------------------------------------------------------------------------------------------------------------LLVVGPKFIRANQEYTLVIS---------------------------------------------------------------------------------------------------------------NFNSQLSKVDLLLKLELSVLNVTKMVDVRRNMNRMINFNMPEDLTAGNYKITIDGQR-GFSFHKEAEL-VYLSKSISGLIQVDKPVFKPGDTVNFRVIVLDTELKPPA-------------------------------RVKSVYVTIRDPQRNVIRKWGVWNISVEVEGEELVSKTFEVKE--------------------------------------------------------------------------------YVLSTFDVQVMPSVIPLEEHQAVNLTIEANYHFGKPVQGVAKVELYLDDDKLKLKKELTVYGKGQVELRFDNFAMQQDVPVKVSFVEQYTNRTVVKQSQITVYR-----------------YAYRVELIKESPQFRPGLPFKCALQFTHHDGTPA----------------------KGISGKVEVSDVRFETTTTSDNDGLIKLELQPSEGTEQLSIHFNAVDGFFFYEDVNK---VETVTDAYIKLELKSPIKRNKLMRFMVTCT------ERMTFFVYYVMSKGNIIDAGFMRP--------------------------------------------------------------------NKQPKYLLQLNATEKMIPRAKILIATVAGRTVVYDFADLAFQEIKPGRQIELSMSGRPGAYVGLAAYDKALLLFNKNHDLFWEDIGQVFDGFHENEFDIFHSLGLFARTLDDILFDSQESWLWKNVSIGRSGSRKLIEVVPDTTTSW--------------------------------------YLTGFSIDPVYGLGIIKKPIQFTT--------- |
| 9 | 1uf2B | 0.11 | 0.09 | 3.41 | 1.11 | MUSTER | | --------------------MDSTGRAYDGAS---EFKSVLVTEGTSHYTPVEVYNILDELKTIKITSTIAEQSVVSRTLSKIGLQDVKKLFDINVIKCGSSLRIVDEPQVTFIVSYAKDIYDKFMCIEHDSAYEPSLTMHRVRV-SMLNDYCAKMISEVPYES--SFVGELPVKSVTLNKLGDRNMDALAE-HLLFEHDVVNAQRENRIFYQRKSAPAVPV-----IFGDLEPAVRERANLYHRYSVPYHQIELALHALANDLLSIQYCHPTVVYNYLSSRAPNFLRVSLKLTSAGIGTLMPRPVVQLLDYDLVYMSPLALN---NLASRL----------LRKISLHLVMQMVTAVQQDLGEVVSVSSNVTN---PASACLVRMNVQGVQTLAVFIAQSMLNP----NISYGMISGTLDCFSNFIYGACLMLFQALIPPSALTARQ--RLDINNRFAYFLIKCHATQATTARLVANQVIPVDAIDQWQSNGRDV------VAIYNNLLPGELVLTNLIQTY--------FRGNTAQQAAEI-----------LIPADQTSYGANETRALSAPY-LFGAPINMLA-PDARLST-YKRDLALPDR-----SPILITTVEGQNSISIENLRHKTGLIRAMY------LNGFVTQPPAWIRNANSNTALLSRPNLLGIYEAILANTYANAVNVY-CDRADIPIEWKLHQSVDPQDLLFGVFGI-VPQYQILNEAVPDFFAGGEDILILQLIRAVYDTLSNKLGRNPADIFHLEEVFKVIEEIVSVQQKIDVRKYFTESMRSGSFSKPRRPVAQRLPNLYSVIMTQADHVYNYMTQLTHIIPITDCFYIVKNSGFVDRGSTGPASSSVYENVLKVVHTIADFDAANALRLQR------------------RRVDNTSYTDSLSDMFNGL-RSISSSEFVRSVNGRDAIKVNMRAKFDLQFITEE--GGYSKPPNVKKLMFSDFLSFLDSHKSDYRPPLLTVPNLGETNSNTLRMRSEAID---------EYFSSYVGAQVPINVVDTRVYTEFSELRNFFTGDVVIRDDPFD--VWDGVKATYIPIGVHGVRLD-PNGDQPPL |
| 10 | 5o9bA | 0.49 | 0.03 | 0.87 | 1.90 | HHsearch | | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------MAHHHHHHSSGLEVLFQGPRRAGTCCANCQTTTTTLWRRNANGDPVCNACGLYYKLHNVNRPLTMKKE------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| (a) | ID1 is the number of template residues identical to query divided by number of aligned residues. |
| (b) | ID2 is the number of template residues identical to query divided by query sequence length. |
| (c) | Cov is equal the number of aligned template residues divided by query sequence length. |
| (d) | Norm. Zscore is the normalized Z-score of the threading alignments. A Normalized Z-score >1 means a good alignment and is highlighted in bold. |
| (e) | Download alignment lists the threading program used to identify the template, and provide the 3D structure of aligned regions of threading templates (threading[1-10].pdb.gz). |
| (f) | Template residues identical to query sequence are highlighted in color. |
|