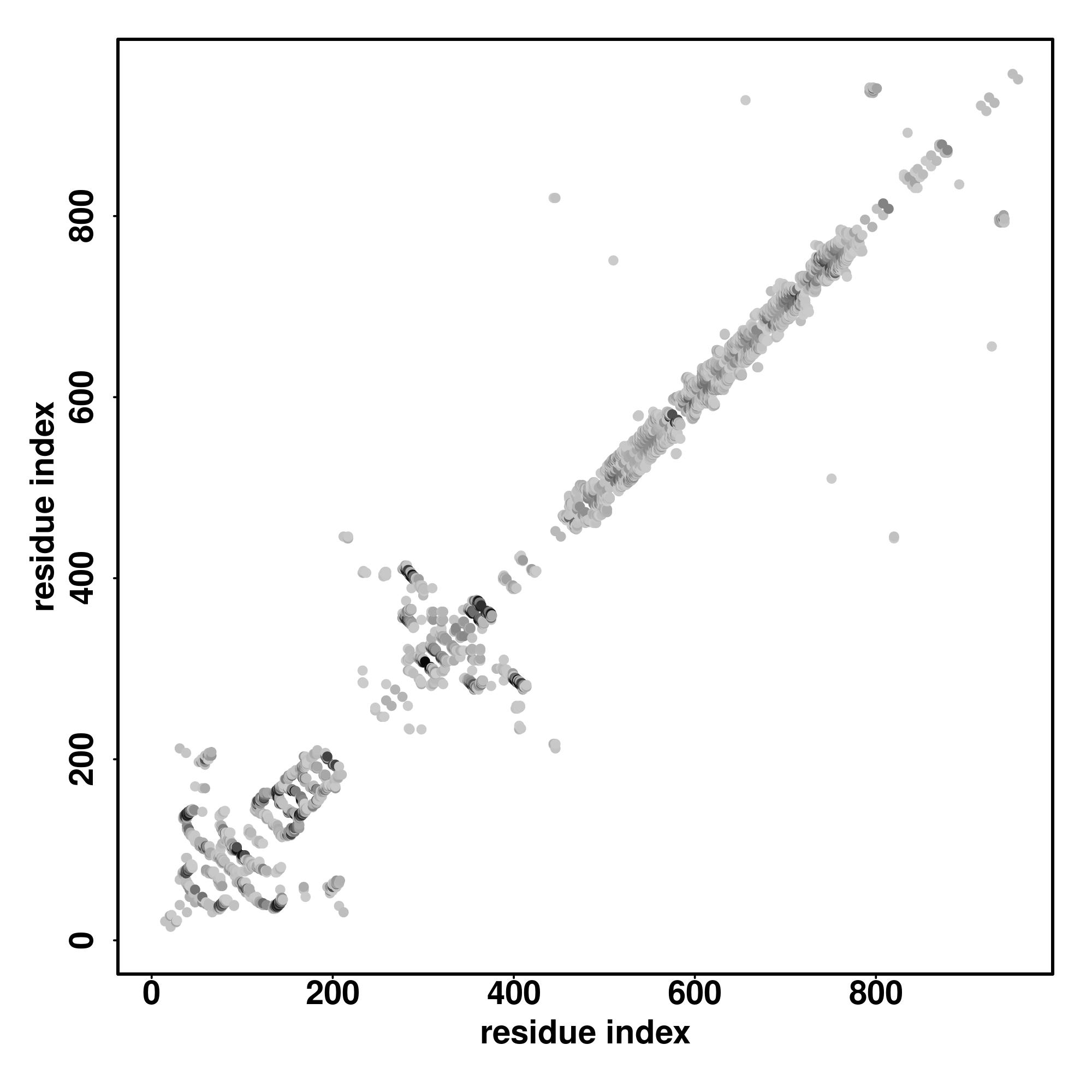
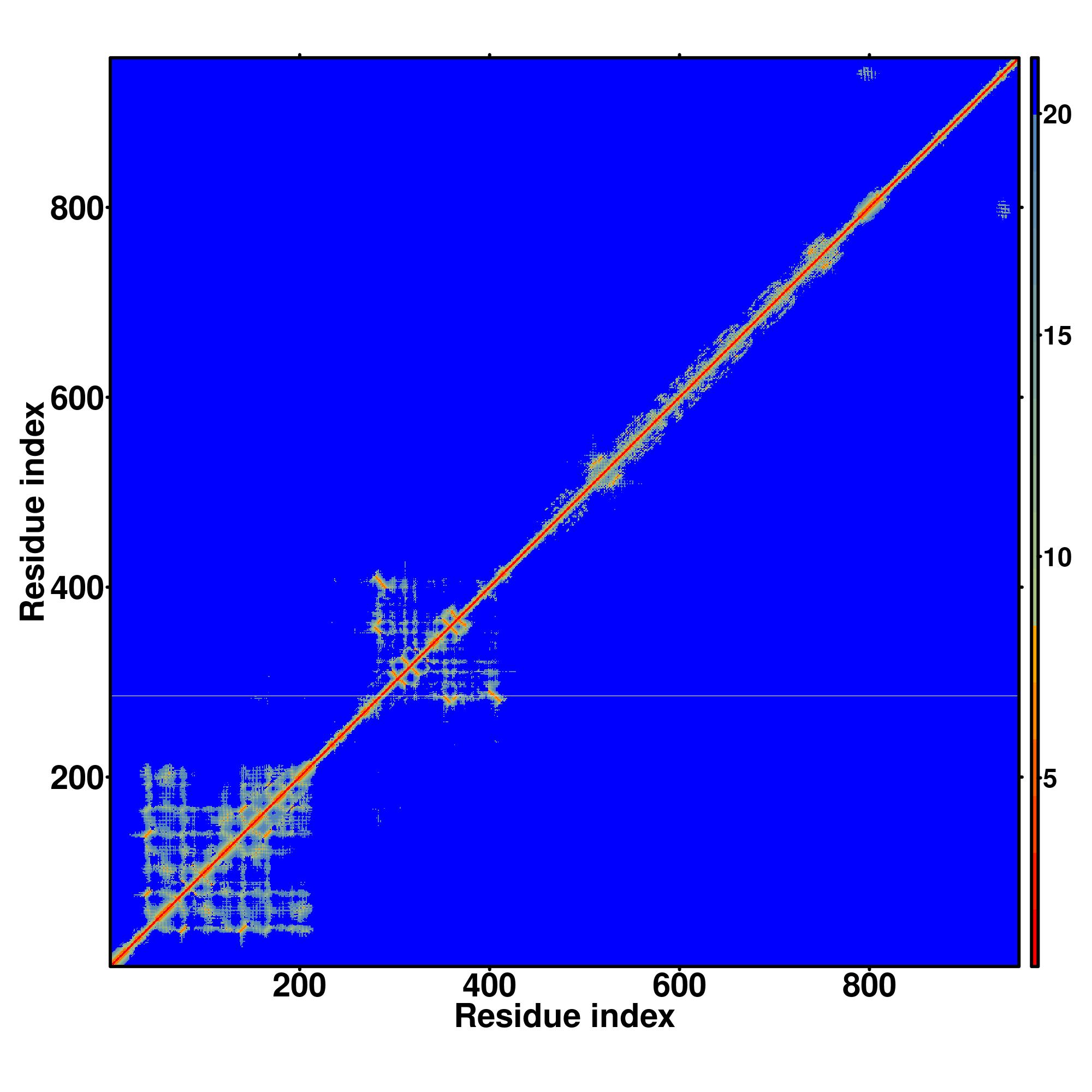
| Rank | PDB
hit | ID1 | ID2 | Cov | Norm.
Zscore | Download
alignment | | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 480 500 520 540 560 580 600 620 640 660 680 700 720 740 760 780
| | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | |
|---|
| SS
Seq | CCCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHCSSSCCCCSSSSSSSSCCCCSSSSSCCCCCCHHHHHHHHHHCHHHCCCCCHHHHHHHHHHHHCCCCCCCCCSSSSSSCCCCCHHHHHHHHHHHHHCCCSSSSSSCCCCCCHHHHHHHCCCCCCCSSSSSCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSSCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
MAHYITFLCMVLVLLLQNSVLAEDGEVRSSCRTAPTDLVFILDGSYSVGPENFEIVKKWLVNITKNFDIGPKFIQVGVVQYSDYPVLEIPLGSYDSGEHLTAAVESILYLGGNTKTGKAIQFALDYLFAKSSRFLTKIAVVLTDGKSQDDVKDAAQAARDSKITLFAIGVGSETEDAELRAIANKPSSTYVFYVEDYIAISKIREVMKQKLCEESVCPTRIPVAARDERGFDILLGLDVNKKVKKRIQLSPKKIKGYEVTSKVDLSELTSNVFPEGLPPSYVFVSTQRFKVKKIWDLWRILTIDGRPQIAVTLNGVDKILLFTTTSVINGSQVVTFANPQVKTLFDEGWHQIRLLVTEQDVTLYIDDQQIENKPLHPVLGILINGQTQIGKYSGKEETVQFDVQKLRIYCDPEQNNRETACEIPGFNGECLNGPSDVGSTPAPCICPPGKPGLQGPKGDPGLPGNPGYPGQPGQDGKPGYQGIAGTPGVPGSPGIQGARGLPGYKGEPGRDGDKGDRGLPGFPGLHGMPGSKGEMGAKGDKGSPGFYGKKGAKGEKGNAGFPGLPGPAGEPGRHGKDGLMGSPGFKGEAGSPGAPGQDGTRGEPGIPGFPGNRGLMGQKGEIGPPGQQGKKGAPGMPGLMGSNGSPGQPGTPGSKGSKGEPGIQGMPGASGLKGEPGATGSPGEPGYMGLPGIQGKKGDKGNQGEKGIQGQKGENGRQGIPGQQGIQGHHGAKGERGEKGEPGVRGAIGSKGESGVDGLMGPAGPKGQPGDPGPQGPPGLDGKPGR |
|---|
| 1 | 3k71G | 0.13 | 0.12 | 4.18 | 0.56 | CEthreader | | ECGRNMYLTGLCFLLGPTQLTQRLPVSRQECPRQEQDIVFLIDGSGSISSRNFATMMNFVRAVISQFQRPST--QFSLMQFSNKFQTHFTFEEFRRSSNPLSLLASVHQLQGFTYTATAIQNVVHRLFHASYGAAAKILIVITDGKKEGDYKDVIPMADAAGIIRYAIGVGLAFSWKELNDIASKPSQEHIFKVEDFDALKDIQNQLKEKIFAIEGTETTSSS----------------------------------------------------------------SFELEMAQEGFSAVFTPDGPVLGAV-GSFTWSGGAFLYPPNMSPTFINMSQENVDMRDSYLGYSTELALWKGVQSLVLGAPRYQHTGKAVIFTQVSRQWRMKAEVTGTQIGSYFGASLCSVDVDSDGSTDLVLIGAPHYYEQTRGGQVSVCPLPRGWRRWWCDAVLYGEQGHPWGRFGAALTVLGDVNGDKLTDVVIGAPGEEENRGAVYLFHGVLGPSISPSHSQRIAGSQLSSRLQYFGQALSGGQDLTQDGLVDLAVGARGQVLLRTRPVLWVGVSMQFIPAEIPRSAFECREQVVSEQTLVQSNICLYIDKRSKNLLGSRDLQSSVTLDLALDPGRLSPRATFQETKNRSLSRVRVLGLKAHCENFNLLLPSCVEDSVTPITLRLNFTLVGKPLLAFRNLRPMLAADAQRYFTASLPFEKNCGADHICQDNLGISFSFPGLKSLLVGSNLELNAEVMVWNDGEDSYGTTITFSHPAGLSYRYVAEGQKQGQLRSL |
| 2 | 3k71G | 0.09 | 0.08 | 2.92 | 1.13 | EigenThreader | | LT---------------------QRLPVSRQECPRQEQFLIDGSGSISSRNFATMMNFVRAVISQFQ---RPSTQFSLMQFSN-KFQTHFTFSVHQL-------------QGFTYTATAIQNVVHRLFHASRRDAAKILIVITDSLDYKDVIPMADAA----GIIRYAIGVQNRNS-------------------WKELND---------------IASKPSQEHIFKVE-----------------DFDALKD------IQNQLKEKIFAIEGTETTSSSS------FELEMAQEGFSAVFTPDGPVLGAVGSFTWSGGAFLYPPNMSPTFINMSNVDMRDSYLGYSTELLVLGAPRYQHVIFTQMKAEVTGTQIGSYFG---ASLCSVDVDSDGSTD-----LVLIGAP-----------HYYEQTRGGQPRGWRRWWCDAVLYGEQGHPWGRFGAVNGDKLTPGEEENRGAVLGPSISPSHSQRIAGSQLSSRLQYLTQDGLVRPVLWPAEIPRSAFECREQVVSEQKRSKNLLGSRDLQPGRLSPRAETKNRLKAHSCVEDSVTPLVGKPLLAFRNLRPMLAADAQRFEKNCGADHICQDNLGPGLKSLNDGEDSYGTVAEGQKQGQLRSLHLGSQGNHLIFRGGAQPKAVLGDENNTPRTSKTTFQEQFTKYLNFSESEEKESHVGQRDLPPVELNQEAVWMDVPQNPSLRPPASDFLAHIQKNPVCSIAGSFSVQEELLSFGWVRQILQKKVDTSVPGQEAFMEKYK------------- |
| 3 | 3hr2A | 0.23 | 0.10 | 3.27 | 0.91 | FFAS-3D | | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GPAGFAGPGADGQGAKGEGDTGVKGDAGPGPAGPAGPGPIGNVGAGPGSRGAAGPGATGFGAAGRVGPGPSGNAGPGPGPVGKEGGKGPRGETGPAGRGEVGPGPGPAGEKGSGADGPAGSGTPGPQGIAGQRGVVGLGQRGKRGFGLGPSGEGKQGPSGASGERGPGPMGPGLAGPGESGREGSGAEGSGRDGAGAKGDRGETGPAGPGAGAGAPGPVGPAGKNGDRGETGPAGPAGPIGPAGARGPAGPQGPRGDGETIGHRGFSGLQGPGSGSGEQGPSGASGPAGPRGGSAGSGKDGLNGLGPIGGPRGRTGDSGPAGPGPGPGPGPPSGGYDFSFLPQPPQEKSQDGGRY- |
| 4 | 4cn8A | 0.24 | 0.06 | 1.94 | 1.58 | CNFpred | | LSTLLKLVIDLACEVC-----------VVDCAG-HADIAFVFDASSSINPNNYQLMKNFMKDIVDRFNKGPDGTQFAVVTFADRATKQFGLKDYSSKADIKGAIDKVTPSIGQTAIGDGLENARLEVFPNR--EVQKVVILLTDGQNNGSPEHESSLLRKEGVVIVAIGVGTGFLKSELINIASSE--EYVFTTSSFDMLSKIMEDVVKLACMSCK------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| 5 | 3k71G | 0.11 | 0.11 | 3.92 | 1.13 | MapAlign | | ECGRNMYLTGLCFLLGPTQLTQRLPVSRQECPRQEQDIVFLIDGSGSISSRNFATMMNFVRAVIS--QFQRPSTQFSLMQFSNKFQTHFTFEEFRRSSNPLSLLASVHQLQGFTYTATAIQNVVHRLFHASYGAAAKILIVITDGKKDSLYKDVIPMADAAGIIRYAIGVGLAFSWKELNDIASKPSQEHIFKVEDFDALKDIQNQLKEKIFGAFLYPPNMSPTFINMSQENVDMRDSYLGYSTELALWKGVQSLVLGAPRYQHTGKAVIFTQVSRQWRMKAEVTGTQIGSYFGASLCSVDVDSDGTDLVLIGAYRGGQVSVCPLPRGWRRWWCYGEGRFGGDVNGDKLTDVVIGANRGAVYLFHGVSHSQRIAGSSRLQYFGQALSGGQDLTQDGLVDLAVGARGQVLLLRTRPVLWVGVSMQFIPAEIPQVVSEQTLVQSNICLYIDKRSKNLLGSRDLQSSVTLDLALDPGRLSPRATFQETKNRSLSRVRVLGLKAHCENFNLLLPSCVEDSVTPITLRLNFTLVGKPLLAFRNLRPMLAADAQRYFTASLPFEKNCGADHICQDNLGISFSFPGLKSLLVGSNLELNAEVMVWNDGEDSYGTTITFSHPAGLSYRYVAEGQKQGQLRSLHLTCDSAPVGSQGTWSTSCRINHLIFRGGAQITFLATFDVSPKAVLGDRLLLTANVSSENNTPRTSKTTFQLELPVKYAVYTVVSSHEQFTKYLNFSESEEKESHVAMHRYQVNNLGQRDLPVSINFWVPVELNQEAVWMDVEVSHPQNPSL |
| 6 | 2pffB | 0.21 | 0.20 | 6.31 | 1.19 | MUSTER | | NDTTLVKTKELIKNYITARIMAKRPFDKKAVGEGNAQLVAIFGGQGN-TDDYFEELRDLYQTVGDLIKFSAETLS-VFTQGLNILEWLENPSNTPDKDYLLSIPISCPLIG---VIQLAHYVVTAKLLGKGATGHSQGLVTAVAIAETDSWESFFVSVRKAITVLFFIGVRNTSLPPSILEDS-EGVPSPMLSISNQEQVQDYVNKTNSHLPAGKQVEISLVNGAKNLVVSGPPQSLYGLNLTLRKAKAPSGLD-----QSRIPFSERKLKFSNRFLPVASPFHS--HLLVPASDLINKDLVKNNVS----FNAKDIQIPVYDTFDGSDLRVLSGSISERIVDCIIRLPVKWETTTQFKATHILDF------------GPGGASGLGVLTHRNKDGTGV-----RVIVAGTLDINPDDDYGFKQGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG--GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG-GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG |
| 7 | 3k71G | 0.11 | 0.11 | 4.01 | 1.01 | SPARKS-K | | ECGRNMYLTGLCFLLGPTQLTQRLPVSRQECPRQEQDIVFLIDGSGSISSRNFATMMNFVRAVISQFQ--RPSTQFSLMQFSNKFQTHFTFEEFRRSSNPLSLLASVHQLQGFTYTATAIQNVVHRLFHASYGDAAKILIVITDGKKEGDYKDVIPMADAAGIIRYAIGVGLRNSWKELNDIASKPSQEHIFKVEDFDALKDIQNQLKEKIFA---IEGTETSSSFELEMAQEGFSAVFTPDGFTWFLYPPNMSPTFINMSQ--ENVDMRDELALWKGVQSLVLGAPRY--QHTGKAVIFTQVSRQWRMKAEVTGTQQVSVCPLPRGWRRWWAALTVLGDV---NGDKLTDVVIGEEENRVYLFHGVGPSISPSHSQRIAGSLQYFGQALSGGQDDGLVDLAVGQVLLLRTRPVLWVGVSMQFIPAEIPRSAFECREQVVSEQTLVQDKRSKNLLGSRDLQSSVTLDLALDPGRLSPRATFQETKNRSLSRVRVLGLKAHCENFNLLLPSCVEDSVTPITLRLNFTLVGKPLLAFRNLRPMLAADAQRYFTASLPFEKNCGADHICQDNLGISFSFPGLKSLLVGSNLELNAEVMVWNDGEDSYGTTITFSHPAGLSYRYVAEGQKQGQLRSLHLTCDSAPVGSQGTWSTSCRINHLIFRGGAQITFLATFDVSPKAVLGDRLLLTANVSSENNTPRTSKTTFQLELPVKYAVYTVVSSHEQFTKYLNFSESEEKESHVAMHRYQVNNLGQRDLPVSINFWVPVELNQEAVWMDVEVSHPQNPSLR |
| 8 | 5es4A | 0.24 | 0.10 | 3.10 | 1.50 | CNFpred | | RNMYLTGLCFLLG------LTQRLPVSRQECPRQEQDIVFLIDGSGSISSRNFATMMNFVRAVISQFQR--PSTQFSLMQFSNKFQTHFTFEEFRRSSNPLSLLASVHQLQGFTYTATAIQNVVHRLFHASYGAAAKILIVITDGKKE-DYKDVIPMADAAGIIRYAIGVGLAFSWKELNDIASKPSQEHIFKVEDFDALKDIQNQLKEKIFAI-MAQEGFSAVFTP--DGPVLGAVGSF----------TWSGGAFLYPSPTFINMSQENVDMRDSYLGYSTELALWK---------------GVQSLVLGAPRTGKAVIFTQVS-RQWRMKAEVTGTQIGSYFG--ASLCSVDVD-TDLVLIGAPH---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| 9 | 3hr2B | 0.29 | 0.21 | 6.50 | 2.00 | HHsearch | | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------SDKGVSAGPGPMGLMGPRGPAGPQGFQGPAGEGAGSRGPAGGERGVVGPQGFGTGLFRGHNGLDGLKGQGA--QGVGEGAGENGTGARGLGERGRVGAGPGARGSDGSVGPVPAGPIGSAGP-GFGAGPPAGPPVGPGNPGANGLTGAATGLGVAGAGLGPRGIGPVGATGPRGLVGSGETGNKERGSPGEAGLSRGLGADGRPPGNRSTGPAGVRGPNDAGRGLMGLG-SGNVGPIGPAGPRGEAGNIGFDGNNGPQGVQGGKGEQGPAGERGLPGEFGPAGPRGERG-PGESGAAGPSGPIGIRGPSGAPGSAGARGEKGETGLRGEPGRDGAAGASGDRGEAGAAGPSGPAGPRGSPGERGEVGPAGPNGFAGPAGEKGTKGPKGENGIVGPTGPSGPNGPPGSRGDGGPPGMTGFRGPRGDQGPVGEIGASGPPGFAGEKGPSGEGTTAGPQGLLGARGEAGAKGERGNIGPTGAAGAPGPHGSVGPAGKHGNRGEPGPAGSVGAVGPRGPSGPQGIRGDKGEPGDQGLPGLAGLHGDQGAPGPVGPAGPRGPAGPSGPIGKVGPAGVRGS |
| 10 | 5bv8A | 0.22 | 0.05 | 1.68 | 1.46 | CNFpred | | --------------------------HDFYCSR-LLDLVFLLDGSSRLSEAEFEVLKAFVVDMMERLRISQKWVRVAVVEYHDSSHAYIGLKDRKRPSELRRIASQVKYAGSVASTSEVLKYTLFQIFSKIDREASRITLLLMASQESRNFVRYVQGLKKKKVIVIPVGIGPHANLKQIRLIEKQAPENKAFVLSSVDELEQQRDEIVSYLCDLAP------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| (a) | ID1 is the number of template residues identical to query divided by number of aligned residues. |
| (b) | ID2 is the number of template residues identical to query divided by query sequence length. |
| (c) | Cov is equal the number of aligned template residues divided by query sequence length. |
| (d) | Norm. Zscore is the normalized Z-score of the threading alignments. A Normalized Z-score >1 means a good alignment and is highlighted in bold. |
| (e) | Download alignment lists the threading program used to identify the template, and provide the 3D structure of aligned regions of threading templates (threading[1-10].pdb.gz). |
| (f) | Template residues identical to query sequence are highlighted in color. |
|