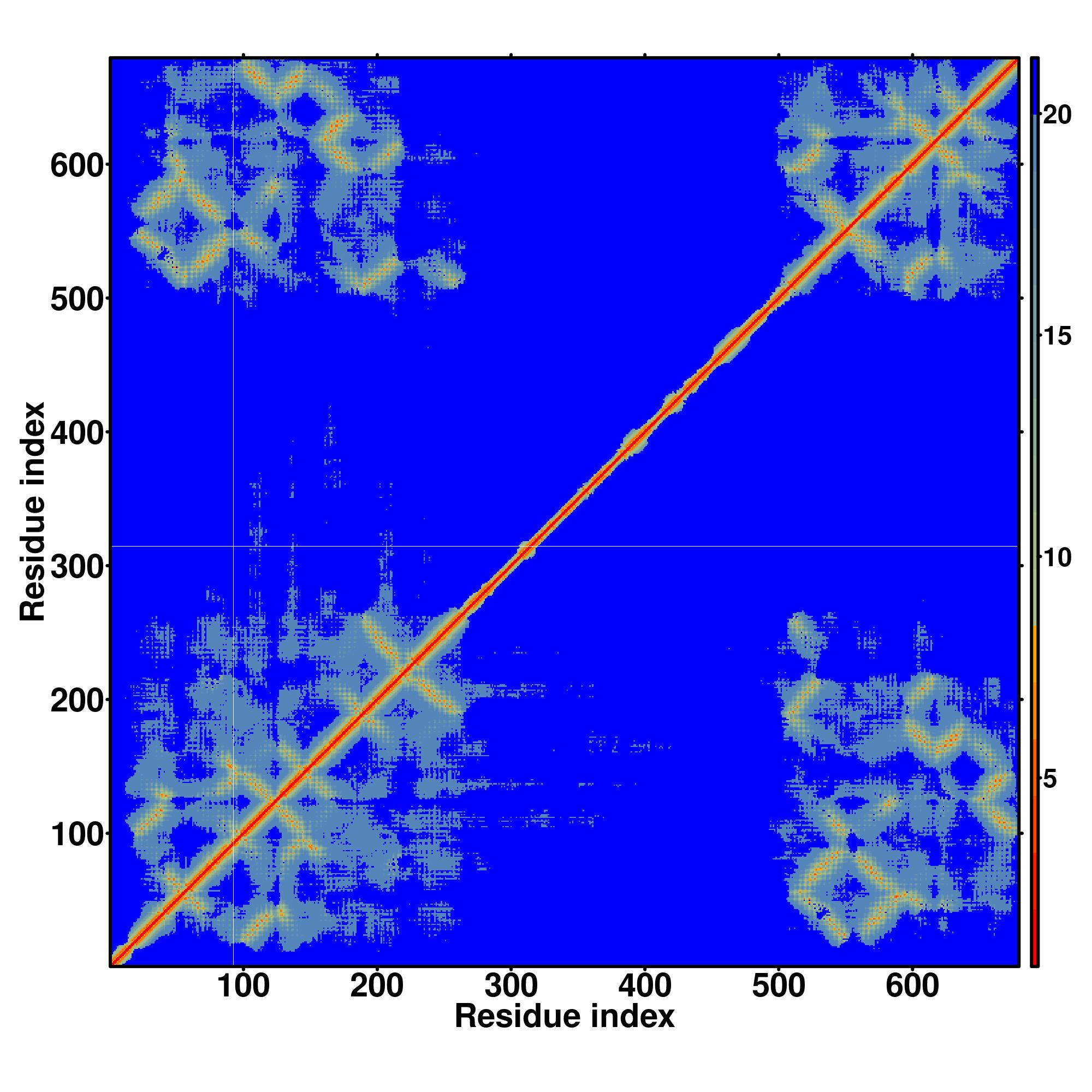
| Rank | PDB
hit | ID1 | ID2 | Cov | Norm.
Zscore | Download
alignment | | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 480
| | | | | | | | | | | | | | | | | | | | | | | | |
|---|
| SS
Seq | CCHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHCHHHHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHCHCHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHCHHHHHHHHHHCCCCCCCCCCCSHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCCCHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCC
MATLITSTTAATAASGPLVDYLWMLILGFIIAFVLAFSVGANDVANSFGTAVGSGVVTLKQACILASIFETVGSVLLGAKVSETIRKGLIDVEMYNSTQGLLMAGSVSAMFGSAVWQLVASFLKLPISGTHCIVGATIGFSLVAKGQEGVKWSELIKIVMSWFVSPLLSGIMSGILFFLVRAFILHKADPVPNGLRALPVFYACTVGINLFSIMYTGAPLLGFDKLPLWGTILISVGCAVFCALIVWFFVCPRMKRKIEREIKCSPSESPLMEKKNSLKEDHEETKLSVGDIESVKAEMGLGDRKGSNGSLEEWYDQDKPEVSLLFQFLQILTACFGSFAHGGNDVSNAIGPLVALYLVYDTGDVSSKVATPIWLLLYGGVGICVGLWVWGRRVIQTMGKDLTPITPSSGFSIELASALTVVIASNIGLPISTTHCKVGSVVSVGWLRSKKAVDWRLFRNIFMAWFVTVPISGVISAAIMAIFRYVILRM |
|---|
| 1 | 6l85A | 0.38 | 0.31 | 9.14 | 1.33 | DEthreader | | ---------------------MTILIIAGILGFIMAFSIGANDVANSMATAVGARAITVRQAALIAMFLEFLGAVMFGSHVSQTIVKGIVEVEKVQ--PVELMYGALSALIAASFWILIATNWGYPVSTTHSIVGGMMGFGLVAVGINGVNWKTFLFIVLSWVVSPVLGGLISFVMFKLISLSVFHTKNPKKSSTVAIPFFISLAIFTMISLFVKK-T--LKQ--P-LSESFLLGIAFSLVTFFVVH---F-AVRKLINEKK------------------------------------------------------DVYDAVENVFKRAQILTSCYVSFSHGANDVANAAGPVAAVMIVASTGVVPKTVEIPFLALLLGGIGISLGVFFLGQKVMETVGEKITTLTNSRGFTVDFSTATTVLLASSLGLPISTTHVVVGAVTGVGFARGLEMVNVGVLKNIVISWLLIVPTVAATSAAVYWVLK-LI--L |
| 2 | 6l85A | 0.38 | 0.31 | 9.19 | 4.64 | SPARKS-K | | ---------------------MTILIIAGILGFIMAFSIGANDVANSMATAVGARAITVRQAALIAMFLEFLGAVMFGSHVSQTIVKGIVEVEKVQ--PVELMYGALSALIAASFWILIATNWGYPVSTTHSIVGGMMGFGLVAVGINGVNWKTFLFIVLSWVVSPVLGGLISFVMFKLISLSVFHTKNPKKSSTVAIPFFISLAIFTMISLFVKKT------LKQPLSESFLLGIAFSLVTFFVVHFAVRKLI-----------NEK-----------------------------------------------KDVYDAVENVFKRAQILTSCYVSFSHGANDVANAAGPVAAVMIVASTGVVPKTVEIPFLALLLGGIGISLGVFFLGQKVMETVGEKITTLTNSRGFTVDFSTATTVLLASSLGLPISTTHVVVGAVTGVGFARGLEMVNVGVLKNIVISWLLIVPTVAATSAAVYWVLKLIL--- |
| 3 | 6l85A | 0.38 | 0.30 | 9.01 | 2.32 | MapAlign | | ---------------------MTILIIAGILGFIMAFSIGANDVANSMATAVGARAITVRQAALIAMFLEFLGAVMFGSHVSQTIVKGIVEVEKV--QPVELMYGALSALIAASFWILIATNWGYPVSTTHSIVGGMMGFGLVAVGINGVNWKTFLFIVLSWVVSPVLGGLISFVMFKLISLSVFHTKNPKKSSTVAIPFFISLAIFTMISLFVKKTL------KQPLSESFLLGIAFSLVTFFVVHFA----VRKLINEKK------------------------------------------------------DVYDAVENVFKRAQILTSCYVSFSHGANDVANAAGPVAAVMIVASTGVVPKTVEIPFLALLLGGIGISLGVFFLGQKVMGEKIT---TLTNSRGFTVDFSTATTVLLASSLGLPISTTHVVVGAVTGVGFARGLEMVNVGVLKNIVISWLLIVPTVAATSAAVYWVLK------ |
| 4 | 6l85A | 0.38 | 0.31 | 9.14 | 1.59 | CEthreader | | ---------------------MTILIIAGILGFIMAFSIGANDVANSMATAVGARAITVRQAALIAMFLEFLGAVMFGSHVSQTIVKGIVEVEKV--QPVELMYGALSALIAASFWILIATNWGYPVSTTHSIVGGMMGFGLVAVGINGVNWKTFLFIVLSWVVSPVLGGLISFVMFKLISLSVFHTKNPKKSSTVAIPFFISLAIFTMISLFVKKTLKQP------LSESFLLGIAFSLVTFFVVHFAVRKLINEK----------------------------------------------------------KDVYDAVENVFKRAQILTSCYVSFSHGANDVANAAGPVAAVMIVASTGVVPKTVEIPFLALLLGGIGISLGVFFLGQKVMETVGEKITTLTNSRGFTVDFSTATTVLLASSLGLPISTTHVVVGAVTGVGFARGLEMVNVGVLKNIVISWLLIVPTVAATSAAVYWVLKLIL--- |
| 5 | 3vvnA | 0.10 | 0.08 | 3.05 | 0.92 | MUSTER | | -----KTTKGVQLLRGDPKKAIVRLSIPMMIGMSVQTLYNL---PESLAAVGL-----FFPVFMGIIALAAGLGVGTSSAIARRIGAR--DKEGA---DNVAVHSLILSLILGVTITITMLP--------------AIDSLFRSMGAE-------AVELAIEYARVLLAGAFIIVFNNVGNGILRGEGD-ANRAMLAMVLGSGLNIVLDPIFIYTLG-----------FGVVGAAYATLLSMVVTSLFIAYWLFVKRDTYVDITLRDFSPSREILKDILRV-------GLPSSLSQLSMSIAMFFLNSVAITAGGENGVAVFTSAWRITMLGIVPILGMAAATSVTGAAYGEVEKLETAYLYA---------IKIAFMIELAVVAFIMLFAPQVAYLFTAQ-ISALRTLPVFLVLTPFGMMTSAMFQGI---GEKSLILTIFRTLVMQ---GFAYIFVHYLRGVWIGIVIGNMVAAIVGFLWGRMRISAL |
| 6 | 6l85A | 0.37 | 0.30 | 9.03 | 8.93 | HHsearch | | ---------------------MTILIIAGILGFIMAFSIGANDVANSMATAVGARAITVRQAALIAMFLEFLGAVMFGSHVSQTIVKGIVEVEKV--QPVELMYGALSALIAASFWILIATNWGYPVSTTHSIVGGMMGFGLVAVGINGVNWKTFLFIVLSWVVSPVLGGLISFVMFKLISLSVFHTKNPKKSSTVAIPFFISLAIFTMISLFVKKTLKQP-L-----SESFLLGIAFSLVTFFVVHFAVRKLINE----------------------------------------------------------KKDVYDAVENVFKRAQILTSCYVSFSHGANDVANAAGPVAAVMIVASTGVVPKTVEIPFLALLLGGIGISLGVFFLGQKVMETVGEKITTLTNSRGFTVDFSTATTVLLASSLGLPISTTHVVVGAVTGVGFARGLEMVNVGVLKNIVISWLLIVPTVAATSAAVYWVLKLIL--- |
| 7 | 6l85A | 0.38 | 0.31 | 9.19 | 3.11 | FFAS-3D | | ---------------------MTILIIAGILGFIMAFSIGANDVANSMATAVGARAITVRQAALIAMFLEFLGAVMFGSHVSQTIVKGIVEVEKVQ--PVELMYGALSALIAASFWILIATNWGYPVSTTHSIVGGMMGFGLVAVGINGVNWKTFLFIVLSWVVSPVLGGLISFVMFKLISLSVFHTKNPKKSSTVAIPFFISLAIFTMISLFVKK------TLKQPLSESFLLGIAFSLVTFFVVHFAVRKL----------------------------------------------------------INEKKDVYDAVENVFKRAQILTSCYVSFSHGANDVANAAGPVAAVMIVASTGVVPKTVEIPFLALLLGGIGISLGVFFLGQKVMETVGEKITTLTNSRGFTVDFSTATTVLLASSLGLPISTTHVVVGAVTGVGFARGLEMVNVGVLKNIVISWLLIVPTVAATSAAVYWVLKLIL--- |
| 8 | 6l85A | 0.37 | 0.30 | 9.02 | 2.83 | EigenThreader | | ---------------------MTILIIAGILGFIMAFSIGANDVANSMATAVGARAITVRQAALIAMFLEFLGAVMFGSHVSQTIVKGIVEV--EKVQPVELMYGALSALIAASFWILIATNWGYPVSTTHSIVGGMMGFGLVAVGINGVNWKTFLFIVLSWVVSPVLGGLISFVMFKLISLSVFHTKNPKKSSTVAIPFFISLAIFTMISLFVKKT-----LKQP-----LSESFLLGIAFSLVTFFVVHFAVRKLI-----------------------------------------------NEKKD-------VYDAVENVFKRAQILTSCYVSFSHGANDVANAAGPVAAVMIVASTGVVPKTVEIPFLALLLGGIGISLGVFFLGQKVMETVGEKITTLTNSRGFTVDFSTATTVLLASSLGLPISTTHVVVGAVTGVGFARGLEMVNVGVLKNIVISWLLIVPTVAATSAAVYWVLK----LI |
| 9 | 3rkoB | 0.12 | 0.09 | 3.30 | 1.69 | CNFpred | | --------------------SAIVGVGSVGLAALVTAFIGVDFFAFNIGFNLVLDGLSLTMLSVVTGVGFLIHMYASWYM----------------GEEGYSRFFAYTNLFIASMVVLVLADN------LLLMYLGWEGVGLCSYLLIGF-DPKNGAAAMKAFVVTRVGDVFLAFALFILYNELG--TLNFREMV-MWATLMLLGGAVGKSAQLPLQTWLADAMAGPTPVSALIHAATMVTAGVYLIARTHGLFLM--------------------------------------------------------------TPEVLHLVGIVGAVTLLLAGFAAVQTDIKRVLAYSTMSQIGYMFLALGVQAWDAAIFHLMTHAFFKALLFLASGSVILACH-HEQNIFKMGGLIPLVYLCFLVGGAALSALPLVTAGFFSKDEILAGAMAN--------HINLMVAGLVGAFMTSLYTFRMIFIVFHGKEQI |
| 10 | 6l85A2 | 0.44 | 0.28 | 8.24 | 1.00 | DEthreader | | ---------------------MTILIIAGILGFIMAFSIGANDVANSMATAVGARAITVRQAALIAMFLEFLGAVMFGSHVSQTIVKGIVEVEKVQ--PVELMYGALSALIAASFWILIATNWGYPVSTTHSIVGGMMGFGLVAVGINGVNWKTFLFIVLSWVVSPVLGGLISFVMFK-------------------------------------------------------------------------------------------------------------------------------------------------------QILTSCYVSFSHGANDVANAAGPVAAVMIVASTGVVPKTVEIPFLALLLGGIGISLGVFFLGQKVMETVGEKITTLTNSRGFTVDFSTATTVLLASSLGLPISTTHVVVGAVTGVGFARGLEMVNVGVLKNIVISWLLIVPTVAATSAAVYWVLK-LI--L |
| (a) | ID1 is the number of template residues identical to query divided by number of aligned residues. |
| (b) | ID2 is the number of template residues identical to query divided by query sequence length. |
| (c) | Cov is equal the number of aligned template residues divided by query sequence length. |
| (d) | Norm. Zscore is the normalized Z-score of the threading alignments. A Normalized Z-score >1 means a good alignment and is highlighted in bold. |
| (e) | Download alignment lists the threading program used to identify the template, and provide the 3D structure of aligned regions of threading templates (threading[1-10].pdb.gz). |
| (f) | Template residues identical to query sequence are highlighted in color. |
|