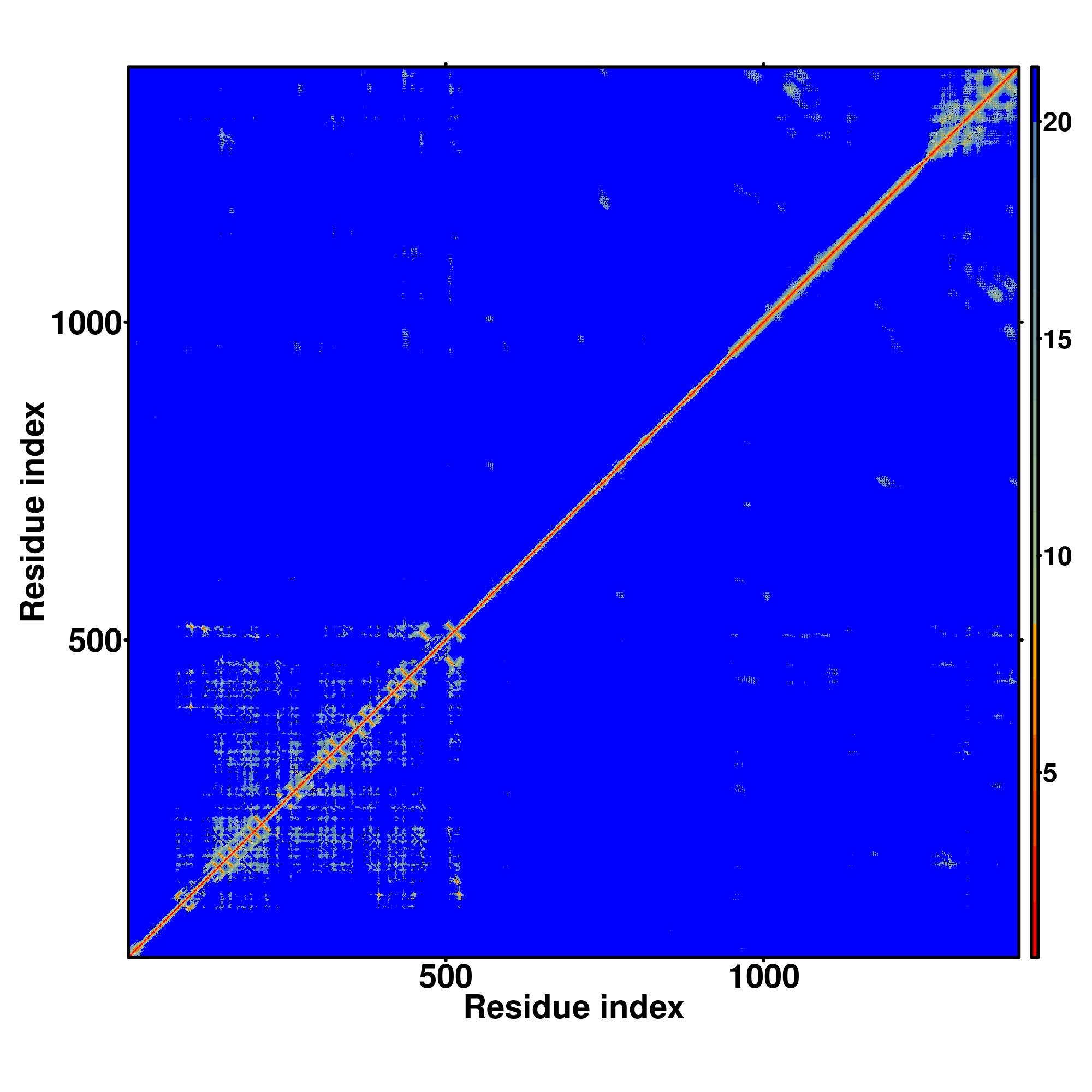
| Rank | PDB
hit | ID1 | ID2 | Cov | Norm.
Zscore | Download
alignment | | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380
| | | | | | | | | | | | | | | | | | | |
|---|
| SS
Seq | CCCCCCCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSSCCCCCCCCSSSSCCCCSSSSCCCCCCCCCCCCCCCCSSSSSSSCCCCCCCCCCSSSSSCCCSSSSSSSCCCCCCSSSSSSSCCCCCCSSSCCCCCCSSSSSSCCCCCCSSSSSSCCCSSSSSSSCCCCCCSSSSSSSSSSCCCCCCCCCCCHHHSSCCCCCHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHCCCCCCSSHHHHHHHHHHHHHHHCCCCSSSSHHHHHHCCCCCHHHHCCCCCCCCCHHHHHHHCCCCCCCCCCCHHHHHCCCHHHHHHHHHHHHHHCCCCCHHHHHHHCC
MASCASIDIEDATQHLRDILKLDRPAGGPSAESPRPSSAYNGDLNGLLVPDPLCSGDSTSANKTGLRTMPPINLQEKQVICLSGDDSSTCIGILAKEVEIVASSDSSISSKARGSNKVKIQPVAKYDWEQKYYYGNLIAVSNSFLAYAIRAANNGSAMVRVISVSTSERTLLKGFTGSVADLAFAHLNSPQLACLDEAGNLFVWRLALVNGKIQEEILVHIRQPEGTPLNHFRQEELLQRLCTQLEGLQSTVTGHVERALETRHEQEQRRLERALAEGQQRGGQLQEQLTQQLSQALSSAVAGRLERSIRDEIKKTVPPCVSRSLEPMAGQLSNSVATKLTAVEGSMKENISKLLKSKNLTDAIARAAADTLQGPMQAAYREAFQS |
|---|
| 1 | 6iamA | 0.15 | 0.10 | 3.48 | 1.10 | SPARKS-K | | ---------------------------------------------------------------------TPVKPNYALKFTLAGHTKAVSSVKFSPNGEWLASSSA--------DKLIKIWGAYDGK-FEKTISGSDVAWSSNLLVSASDDK-----TLKIWDVSSGCLKTLKGHSNYVFCCNFNP-QSNLIVSGSFDESVRIWDV--KTGKCLKTLVSAVHFNRDGSLYDGLCRIWDTASGQCLKTLIDKFSPNGKYILAATLDNTLKLWDYSKGKCLKTYTGHKNEKYVTGGKWIVSGSDNLV--YIWNLQTKEI----VQKLQGHTDV-VISTACH---PTENENDKTIKLWKSDC--------------------------- |
| 2 | 2ovpB | 0.12 | 0.09 | 3.03 | 2.97 | CNFpred | | -------------------------------------------------------------------------------KVLKGHDHVITCLQFCGNRIVSGSDDN----------TLKVWSAVTGKCLRTL-GVWSSQMRDNIIISGSTD-----RTLKVWNAETECIHTLYGHTSTVRCMHLHE---KRVVSGSRDATLRVWDIETGQCLHVL-AVRCVQYDVVSGAYDFMVKVWDPETETCLH-RVYSLQFDGIHVVSGSLDTSIRVWDVETGNCIHTLTGHQSLTSGMELNILVSGNAD-STVKIWDIKT----GQCLQTLQGPNKHAVTCLQFNKNFVITSSDDGTVKLWDLKTGEFIRNLVTLES-GGSGGVVWRIRASN |
| 3 | 5mzhA | 0.08 | 0.07 | 2.64 | 0.31 | CEthreader | | PLTNCAFNKSGDRFITGSYDRTCKVWNTFTGEEVFTLEGHKNVVYAIAFNNPYGDKIVTGSFDKTCKLWDAYTGQLYYTLKGHQTEIVCLSFNPQSTIIATGSMDNTAKLWDVETGQERATLAGHRAE----IVSLGFNTGGDLIVTGSF-----DHDSRLWDVRTGCVHVLSGHRGEVSSTQFNY-AGTLVVSGSIDCTSRLWDVRS-------GRCLSVKQGHTDEVLD------------------VAFDAAGTKMVSASADGSARLYHTLTGVCQHTLVGHEGEISNPQGTRLITASSDK-TCRLWDCDTGECLQVLEGHTD-----EIFSCAFNYEGDFTGSKDNTCRIWKALT--------------------------- |
| 4 | 6eoqA | 0.07 | 0.06 | 2.70 | 0.63 | EigenThreader | | HQDDPVATFVIQEEF------DRFPTASWEGSLPALEERKTDSY--RYPRTGSKNPSQGKELVQPFSSLFPKRDGKYAWAMFLDRPQQWQLVLLPQPYVVYEEVTNVWINVPEFKCPIKEEIALTSGEWEVLARHGSVNEETKLVYFQGTKDTPLEHHLYVVSYEAAGEIVRLTTPGFSHSCSMS-QNFDMFVSSSVSTPPHVYKLSGPLHKQPRFWASMME-----ADYVPPEIFHFHTRSDV-RLYGMIYKPHALTVLFVYKGIKYLRLNTLASLGYAVVVIDGRGLKNQMGQVAIHGWSYGGFLSLMGLIHKPQFKVAIAGAPVTVWMAYDLILHGFLDENVHFKPYQLQIYPNERIRCPESGEHYEVTLLHFLQEYLH---- |
| 5 | 7abhE | 0.14 | 0.11 | 3.92 | 0.47 | FFAS-3D | | --------AQRKQQMAEEMVEAAGELAAEMAAAFLNENLPESIFGAPKAGNGQWASVIRVMNPIQGNTLDLVQLEQNEAAFFSNTGEDWYVLVGVGFVYTYKLVNNGEKLEFLHKTPVEPAAIAPFQ------GRVLIGVGKLLRVYDLGKKKHIANYISGIQT-IGHRVIVSDVQESFIWVRYSLLDYDTVAGADKFGNICVVRLPPNTNKAEVIMNYHV----GETVLSLQKTTLIPGGSESL--VYTTLSGGIGILVPFTSHEDHDFFQH-----------VEMHLRSEHPP-----LCG-------------RDHLSFRSYYFPVKNV----------IDGDLCEQFNSMEKQKNVSEELDRTPPEVSKKLEDIRTRYAF-- |
| 6 | 5mzhA | 0.12 | 0.10 | 3.68 | 1.09 | SPARKS-K | | ------------KLRKFLLRYYPPGIILQYERVMKQKPIDLLDLTPDVDVEVLLSQQEPLISENRRPALRDKMLEFTLFKVLRAHILPLTNCAFNKSGDRFITGSY--------DRTCKVWNTFTGE-EVFTLEGYAIAFNGDKIVTGSFDK-----TCKLWDAYTGLYYTLKGHQTEIVCLSFNPQ-STIIATGSMDNTAKLWDV--ETGQERATLIVSLGFNTGGDLFDHDSRLWDVRTGQCGEVSSTQFNYAGTLVVSGSIDCTSRLWDVRSGRCLSVKQGHTDEVLDAAGTKMVSASADG-SARLYHTLTGVC----QHTLVGH-EGEISKVAFN---PQGTRSDKTCRLWDCDTGEC------LQVLEGHTDEIFSCAFNY |
| 7 | 2ynpA | 0.09 | 0.06 | 2.35 | 2.91 | CNFpred | | ---------------------------------------------------------------------------LDIKKTFSNRSRVKGIDFHPTEPWVLTTLY---------SGRVELWNYEVEVRSIQV-PVRAGKFIANWIIVGSDD-----FRIRVFNYNTEKVVDFEAHPDYIRSIAVHPT-KPYVLSGSDDLTVKLWNWEN-----NWALEQTFEG--------HEHFVMCVAFNP----------KDPSTFASGCLDRTVKVWSLGQSTPNFTLTTGQRGVNY-DKPYMITASDD-LTIKIWDYQT----KSCVATLEGHM-SNVSFAVFHPT-IISGSEDGTLKIWNSSTYKVE------KTLNVGLERSWCIATHP |
| 8 | 6qp7A | 0.07 | 0.05 | 2.13 | 0.83 | DEthreader | | FNCGKLY-Y--RT--F---------------------------YVGAMDRV-----------NR-VINLEPTRDDVSNHVVIQSMDGLYVCGTNHNPKDYVIYANLTYLPRSEYVIGVGLYSGIRTLKYDSWLDKPNFVGSFDYFFFRETAVIGKAVYSRIARVCKTYLKARLNCFYFNEIQSVYQLDRFFATFTTSTIGSAVCSFEIQAFNKAVNHNNPVYY-----KRD--LV-FTK-L---------VVDKIIVYYVGT---NLGRIYKIVQY---------------LLDIFE--ISQTRKSLY-----RIKQIDLAMCNR---------DPYCGWDKEANTCRPYELLLQDVA--NE--TS-DI-CDSSVL-CHLGGSL-- |
| 9 | 1pevA | 0.05 | 0.05 | 2.20 | 0.42 | MapAlign | | ALFPSLPRTARGTAVVLGNTPAGDKIQYCNGTSVYTVPVGSLTDTEIYTEHSHQTTVAKTSPSGYY----CASGDVHGNVRIWDTHILKTTIVFSGPVSKRIAAVGEGRERFGHVFLFDTGTSNGN-LTGQARAMNSVDKPSFRIISGSDD-----NTVAIFEGPPFKKSTFGEHTKFVHSVRYNPD-GSLFASTGGDGTIVLYNGV------DGTKTGVFAHSGSVTIPVGTRIEDQQLGIIWTKQALVSISANGFINFVNPELGSIDQVNSWNRVFPDVHATMITGIKTTSKGDLFTVSWDHLKVVPAG--GSGVDSSKAVANKLS--SQPLGLAVSADGDIAVAACYSCVALSNDKQFVAVGGQDSKVHVYKLSGASVSEVKT |
| 10 | 4wjsA | 0.14 | 0.13 | 4.50 | 0.86 | MUSTER | | KDLIIDQYPNDLLSLLQKH--VTNPFETTITLSAEPQAIFKVHAVSRLAHRIPGHGQPILSCQFSPVSSSRATGSGDNTARIWDTDSGTPKFTLKGHTGWVLGVSWSPDGKY-MDTTVRVWDPESGKQVNQEFRGLALAWQTARLASASK-----DCTVRIWLVNTGTEHVLSGHKGSVSCVKWGGTD--LIYTGSHDRSVRVWDAVKGGHWVNHIALLRTAYHDHTKEVPGTEEERRAKAKERFEKAAKIKGKVAERLVSASDDFTMYLWDNNGSKPVARLLGHQNKVNSPDGTLIASAGWDNSTKLWNARDGK-----FIKNLRGHVA-PVYQCAWS---ADSRLKDCTLKVWNVRTGKL------AMDLPGHEDEVYAVDWAA |
| (a) | ID1 is the number of template residues identical to query divided by number of aligned residues. |
| (b) | ID2 is the number of template residues identical to query divided by query sequence length. |
| (c) | Cov is equal the number of aligned template residues divided by query sequence length. |
| (d) | Norm. Zscore is the normalized Z-score of the threading alignments. A Normalized Z-score >1 means a good alignment and is highlighted in bold. |
| (e) | Download alignment lists the threading program used to identify the template, and provide the 3D structure of aligned regions of threading templates (threading[1-10].pdb.gz). |
| (f) | Template residues identical to query sequence are highlighted in color. |
|