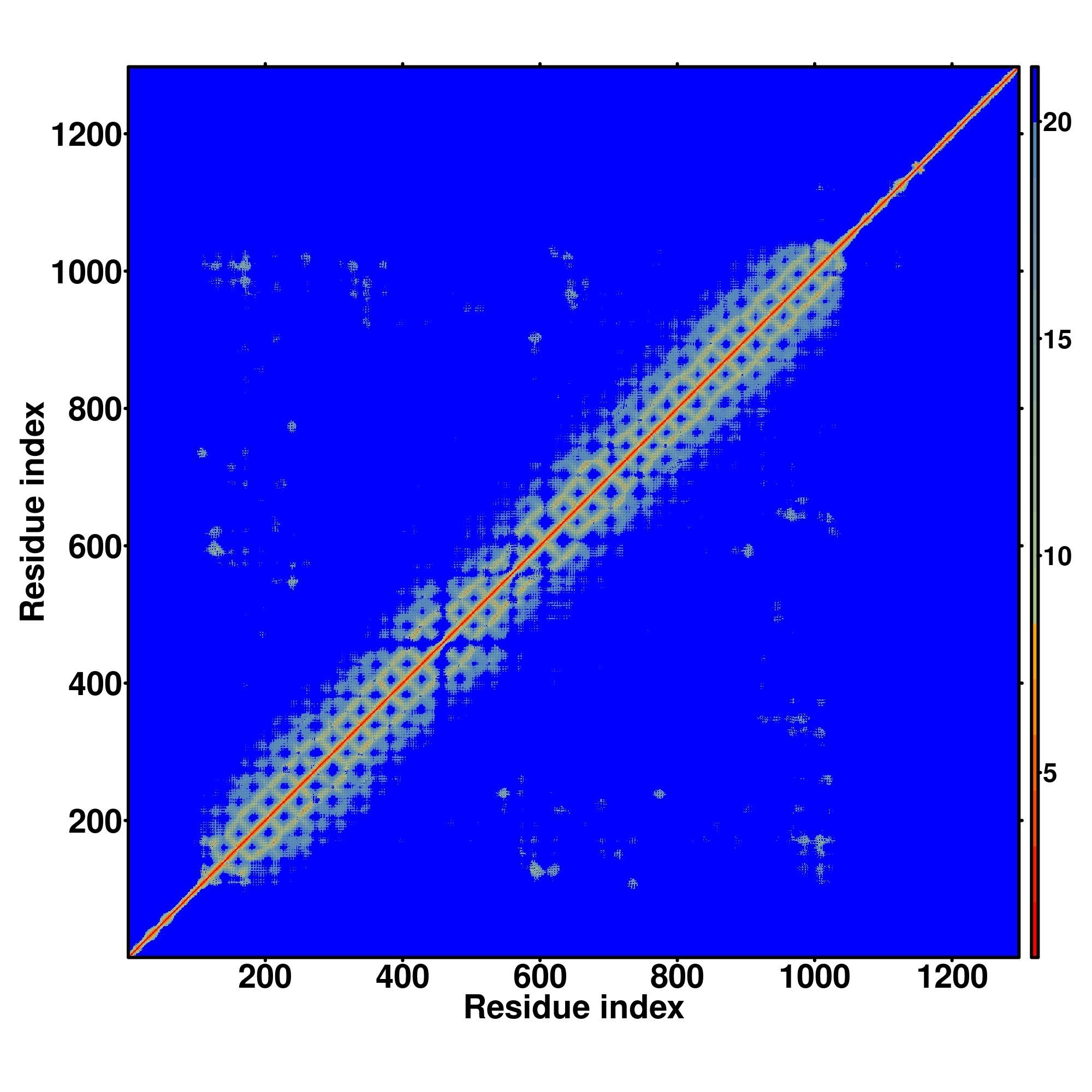
| Rank | PDB
hit | ID1 | ID2 | Cov | Norm.
Zscore | Download
alignment | | 20 40 60 80 100 120 140 160 180
| | | | | | | | | |
|---|
| SS
Seq | CCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHCCCC
QETRLGFFTTYFLPLANTLKSKAMDLAQAGSTVESKIYDTLQWQMWTLLPGFCTRPTDVAISFKGLARTLGMAISERPDLRVTVCQALRTLITKGCQAEADRAEVSRFAKNFLPILFNLYGQPVAAGDTPAPRRAVLETIRTYLTITDTQLVNSLLEKASEKVLDPASSDFTRLSVLDLVVALAPCA |
|---|
| 1 | 6fvbA | 0.09 | 0.08 | 3.10 | 1.17 | DEthreader | | YDPQVCDFIKLSVSHFELISNHENFKK-------FDIYEKFIKCLGKLYFNLVGSANFILPCSTQILITYTRLIFKWEQTAIRGLLILKRVINFIKKIDASINKINTLNELITRLVDTLEWYRRPTELNYQIRPCAENVFQDLNT-F-S--EL-LVPYLLKKIEASKLDFLRKDAIYASFQLSASAV |
| 2 | 5cwfA | 0.09 | 0.08 | 3.06 | 0.83 | SPARKS-K | | -------MSDEMKKVMEALKKAVELAKKNNDDEVAREIERAAKEIVEALREN--NSDEMAKVMLALAKAVLLAAKNNDEVAREIARAAAEIVEALRENDEMAKVMLALAKAVLLAAKN---------NDDEVAREIARAAAEIVEALREKKMLELAKRVLDAAKNNDDETAREIARQAAEEVEAD-- |
| 3 | 1qgkA | 0.06 | 0.06 | 2.57 | 0.61 | MapAlign | | --STEHMKESTLEAIGYICLTAIIQGMSNNVKLAATNALLNSVAALQNLVKIMSLYQYMETYMPALFAITIEAMKDIDEVALQGIEFWSNVCDEEMDLAISKFYAKGALQYLVPILTQTLTKQDDDDDDWNPCKAAGVCLMLLATCC-EDDIVPHVLPFIKEH-IKNPDWRYRDAAVMAFGCILEGP |
| 4 | 6tc0C3 | 0.09 | 0.09 | 3.43 | 0.41 | CEthreader | | FQTASERVEAEGLAALHSLTACLSCSVLRAEDLLGSFLSNILQDCRHHLCEPDASARACEHLTSNVLPLLLEQFHKHSNQRRTILEMILGFLKLQQKWSRDERPLSSFKDQLCSLVFMALT-----DPSTQLQLVGIRTLTVLGAQLSAEDLELAVGHLYRLTFLEEDSQSCRVAALEASGTLATLY |
| 5 | 4uaeA3 | 0.13 | 0.11 | 3.90 | 0.60 | MUSTER | | TDEQTQVVLNC------DALSHFPALLTHPK---EKINKEAVWFLSNIT----GNQQQVQAVIDALVPMIIHLLDKDFGTQKEAAWAISNLTISG--RKDQVAYLIQQ---VIPPFCNLL-----TVKDAQVVQVVLDGLSNILKMAEETIGNLIEECGLEKIEQNHENEDIYKLAYEIIDQFFSS- |
| 6 | 6n1zA | 0.18 | 0.17 | 5.59 | 0.71 | HHsearch | | RIAAQDESAAALAAAATRHLQEAEQTKNSGTIHEHGFLTNVILAALWAASRFTVAMS--PELIQQFLQATVSGLHEPPSVRISAVRAIWGYCDQLK---VSTHVLQPFLPSILDGLIHLAAQFSNDTDDNATMQNGGECLRAYVSVTLEQVAQWYVMQVVSQLLDPRTSEFTAAFVGRLVSTLISKA |
| 7 | 6tc0C3 | 0.11 | 0.10 | 3.68 | 0.92 | FFAS-3D | | ----EAEGLAALHSLTACLSCSVLRADAEDLSFLSNILQDCRHHLAKLLQAAAGSARACEHLTSNVLPLLLEQFHKHSNQRRTILEMILGFLKLQQKEDRDERPLSSFKDQLCSLVFMALT-----DPSTQLQLVGIRTLTVLGAQLSAEDLELAVGHLYRLTFLEEDSQSCRVAALEASGTLA--- |
| 8 | 3fgaB1 | 0.07 | 0.06 | 2.60 | 0.72 | EigenThreader | | PADQEKLFIQKLRQCCVLF----------DFV---SKWKEVKRAALSEMVEYITRNVITEPIYPEVVHMFAVNMFAAWPHLQLVYEFFLRFLESPDFQ---PNIAKKYIDKFVLQLLELFDSED-----PRERDFLKTTLHRIYGKFLRAYIRKQINNIFYRFIYETEHHN---GIAELLEILGSII |
| 9 | 2bkuB | 0.12 | 0.11 | 3.81 | 0.86 | CNFpred | | ----AETSASISTFVMDKLGQTMSVDENQLTLEDAQSLQELQSNILTVLAAVIRKPSSVEPVADMLMGLFFRLLEKSAFIEDDVFYAISALAASLG------KGFEKYLETFSPYLLKALNQV-----DSPVSITAVGFIADISNSLFRRYSDAMMNVLAQMISNPNARRELKPAVLSVFGDIASNI |
| 10 | 3ea5B | 0.08 | 0.07 | 2.76 | 1.17 | DEthreader | | IKMERNYLMQVVCEATQ-AE-----------------DIEVQAAAFGCLCKIMSKYTFMKPYMEALYALTIATMKSNDKVASMTVEFWSTICEEEIILLQSYNFALSSIKDVVPNLLNLL-TRQNEDDDWNVSMSAGACLQLFAQNCGN-HILEPVLEFVEQNITA-DNWRNREAAVMAFGSIMDGP |
| (a) | ID1 is the number of template residues identical to query divided by number of aligned residues. |
| (b) | ID2 is the number of template residues identical to query divided by query sequence length. |
| (c) | Cov is equal the number of aligned template residues divided by query sequence length. |
| (d) | Norm. Zscore is the normalized Z-score of the threading alignments. A Normalized Z-score >1 means a good alignment and is highlighted in bold. |
| (e) | Download alignment lists the threading program used to identify the template, and provide the 3D structure of aligned regions of threading templates (threading[1-10].pdb.gz). |
| (f) | Template residues identical to query sequence are highlighted in color. |
|