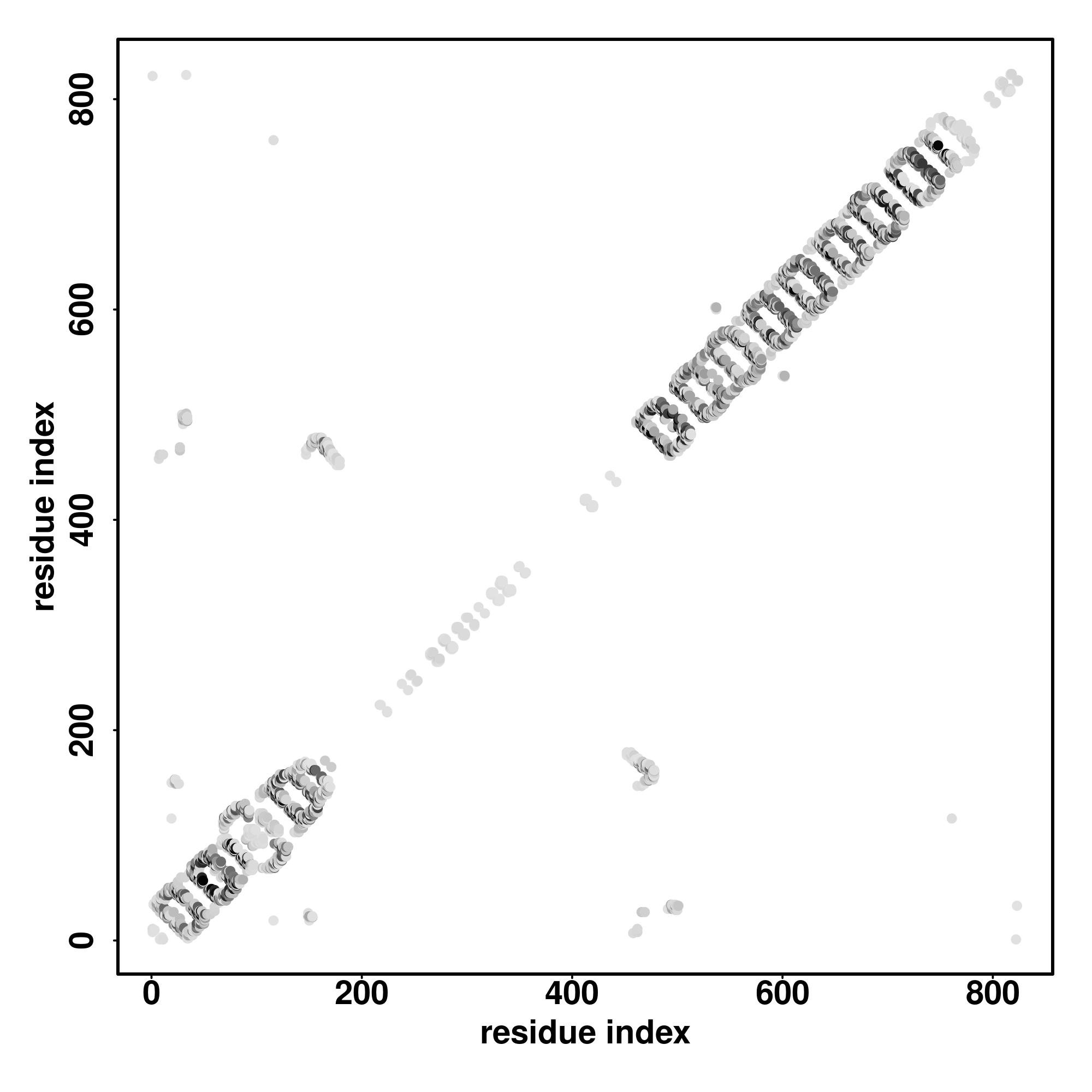
| Rank | PDB
hit | ID1 | ID2 | Cov | Norm.
Zscore | Download
alignment | | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380
| | | | | | | | | | | | | | | | | | | |
|---|
| SS
Seq | CCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCCC
TITPQIQAFNLQKAAAEGLMSLLREMGKGYLALCSYNCKEAINILSHLPSHHYNTGWVLCQIGRAYFELSEYMQAERIFSEVRRIENYRVEGMEIYSTTLWHLQKDVALSVLSKDLTDMDKNSPEAWCAAGNCFSLQREHDIAIKFFQRAIQVDPNYAYAYTLLGHEFVLTEELDKALACFRNAIRVNPRHYNAWYGLGMIYYKQEKFSLAEMHFQKALDINPQSSVLLCHIGVVQHALKKSEKALDTLNKAIVIDPKNPLCKFHRASVLFANEKYKSALQELEELKQIVPKESLVYFLIGKVYKKLGQTHLALMNFSWAMDLDPKGANNQIKEAIDKRYLPDDEEPITQEEQIMGTDESQESSMTDADDTQLHAAESDEF |
|---|
| 1 | 4ui9J | 0.19 | 0.16 | 5.24 | 1.33 | DEthreader | | --------------LCNEEQELLRFLFENKLKKYNKPSETVIPE-SVNGLEK--NLDVVVSLAERHYYNCDFKMCYKLTSVVMEKDPFHASCLPVHIGTLVELNKANELFYLSHKLVDLYPSNPVSWFAVGCYYLMVGKNEHARRYLSKATTLEKTYGPAWIAYGHSFAVESEHDQAMAAYFTAAQLMKGCHLPMLYIGLEYGLTNNSKLAERFFSQALSIAPEDPFVMHEVGVVAFQNGEWKTAEKWFLDALEKIKVWEPLLNNLGHVCRKLKKYAEALDYHRQALVLIPQNASTYSAIGYIHSLMGNFENAVDYFHTALG-LRRDD-TFSVTMLGHCIEM--------------------------------------- |
| 2 | 1w3bA | 0.14 | 0.14 | 4.82 | 2.00 | SPARKS-K | | MELAHREYQAAAERHCMQLWRQGVLLLLSSIHFQCRRLDRSAHFSTLAIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAYVSALQYNPDLYCVRSDLGNLLKALGRLEEAKACYLKAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVAAYLRALSLSPNHAVVHGNLACVYYEQGLIDLAIDTYRRAIELQPHFPDAYCNLANALKEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQD---- |
| 3 | 2xm6A | 0.11 | 0.11 | 3.86 | 0.71 | MapAlign | | ---KDLTQAMDWFRRAAEQGYTPAEYVLGLRYMNGQDYAQAVIWYKKAA--LKGLPQAQQNLGVMYHGVKVKAESVKWFRLAAEQ--GRDSGQQSMGDAYFDGRDYVMAREWYSKAAEQ--GNVWSCNQLGYMYSGVRNDAISAQWYRKSATSG--DELGQLHLADMYYIGVDYTQSRVLFSQSAEQ--GNSIAQFRLGYILELAKEPLKALEWYRKSAEQ--GNSDGQYYLAHLYDGVKNREQAISWYTKSAEQG--DATAQANLGAIYFRLEEHKKAVEWFRKAAAKG--EKAAQFNLGNALLKGKDEQQAAIWMRKAAEQGLSAAQVQLGEIYYYLGVERDYVQAWAWFDTASFGTENRNITEKKLTAKQLQQAELLS |
| 4 | 1w3bA | 0.14 | 0.14 | 4.75 | 0.46 | CEthreader | | AGDFEAAERHCMQLWRQEPDNTGVLLLLSSIHFQCRRLDRSAHFSTLAIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAYVSALQYNPDLYCVRSDLGNLLKALGRLEEAKACYLKAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVAAYLRALSLSPNHAVVHGNLACVYYEQGLIDLAIDTYRRAIELQPHFPDAYCNLANALKEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQD---- |
| 5 | 1w3bA | 0.15 | 0.14 | 4.87 | 1.79 | MUSTER | | AERHCMQLWRQEPDN----TGVLLLLSSIHFQCR--RLDRSAHFSTLAIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAYVSALQYNPDLYCVRSDLGNLLKALGRLEEAKACYLKAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVAAYLRALSLSPNHAVVHGNLACVYYEQGLIDLAIDTYRRAIELQPHFPDAYCNLANALKEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQD---- |
| 6 | 6af0A | 0.12 | 0.12 | 4.33 | 0.87 | HHsearch | | SLNDAAEQLRNALKSFEEAIRMLAVMGKARALFSLGRYPESLAAYQDVVAKMPDDPDPRIGIGCCFWQLGFKDDAKIAWERCLEINPDSKHANILLGLYYLDASLYKKATEYTQKSFKLDKNLPLTCATFAGYFLSRKQFGNVDALAHKAIQYTDVNADGWYLLARKEHYDGNLERASDYYRRADDARGGYLPAKFGAAQLSVLKNDLGEAKLRLEKMIQ-HSKNYEAMILLGTLYAESAEPDKALQCLQQVEQLEIDQPQLLNNIGCFYSQEGKHRLATEFFQAALDSCARLTTIPFNLGRSYEYEGDIDKAIETYEQLLSRHSDYTDARTRLAYIKLRRNPNKEGPDAVAKLYQENPSDLEVRGLYGWFLSKVNSKKPE |
| 7 | 1w3bA | 0.15 | 0.15 | 4.93 | 3.11 | FFAS-3D | | AERHCMQLWRQEPGVLLLLSSIHFQCR---------RLDRSAHFSTLAIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAYVSALQYNPDLYCVRSDLGNLLKALGRLEEAKACYLKAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVAAYLRALSLSPNHAVVHGNLACVYYEQGLIDLAIDTYRRAIELQPHFPDAYCNLANALKEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQD---- |
| 8 | 6vbu4 | 0.17 | 0.17 | 5.59 | 1.00 | EigenThreader | | KQNWLIHLYYIQKAVIKEQLQETHIYVQALIFRLEGNIQESLRLFQMCAFLSPQCADNLKQVARSLFLLGKHKAAIEVYNEAAKLNQKDWEICHNLGVCYIYLKQFDKAQDQLHNALHLNR-HDLTYIMLGKIFLLKGDLDKAIEIYKKAVEFSPENTELLTTLGLLYLQLGIYQKAFEHLGNTLTYDPTNYKAILAAGSMMQTHGDFDVALTKYKVVACAVIESPPLWNNIGMCFFGKKKYVAAISCLKRANYLAPLDWKILYNLGLVHLTMQQYASAFHFLSAAINFQPKMGELYMLLAVALTNLEDSENAKRAYEEAVRLDKCNLVNLNYAVLLYNQGEKRDALAQYQEMEK-----KVSSSLEFDPEMVEVAQKL-- |
| 9 | 4ui9F | 0.99 | 0.87 | 24.26 | 2.09 | CNFpred | | -------AFNLQKAAAEGLMSLLREMGKGYLSLCSYNCKESINILSHLPSHHYNTGWVLCQIGRAYFELSEYMQAERIFSEVRRIENYRVEGMEIYSTTLWHLQKDVALSVLSKDLTDMDKNSPEAWCAAGNCFSLQREHDIAIKFFQRAIQVDPNYAYAYTLLGHEFVLTEELDKALACFRNAIRVNPRHYNAWYGLGMIYYKQEKFSLAEMHFQKALDINPQSSVLLCHIGVVQHALKKSEKALDTLNKAIVIDPKNPLCKFHRASVLFANEKYKSALQELEELKQIVPKESLVYFLIGKVYKKLGQTHLALMNFSWAMDLDPKGANNQIKEAIDKR------------------------------------------ |
| 10 | 6tntJ | 0.19 | 0.16 | 5.10 | 1.33 | DEthreader | | --------------LCNEEQELLRFLFENKLKKYNKPSET-VIP-ESVDGLQE-NLDVVVSLAERHYYNCDFKMCYKLTSVVMEKDPFHASCLPVHIGTLVELNKANELFYLSHKLVDLYPSNPVSWFAVGCYYLMVGKNEHARRYLSKATTLEKTYGPAWIAYGHSFAVESEHDQAMAAYFTAAQLMKGCHLPMLYIGLEYGLTNNSKLAERFFSQALSIAPEDPFVMHEVGVVAFQNGEWKTAEKWFLDALEKIKVWEPLLNNLGHVCRKLKKYAEALDYHRQALVLIPQNASTYSAIGYIHSLMGNFENAVDYFHTALG-LRRDD-TFSVTMLGHCIEM--------------------------------------- |
| (a) | ID1 is the number of template residues identical to query divided by number of aligned residues. |
| (b) | ID2 is the number of template residues identical to query divided by query sequence length. |
| (c) | Cov is equal the number of aligned template residues divided by query sequence length. |
| (d) | Norm. Zscore is the normalized Z-score of the threading alignments. A Normalized Z-score >1 means a good alignment and is highlighted in bold. |
| (e) | Download alignment lists the threading program used to identify the template, and provide the 3D structure of aligned regions of threading templates (threading[1-10].pdb.gz). |
| (f) | Template residues identical to query sequence are highlighted in color. |
|