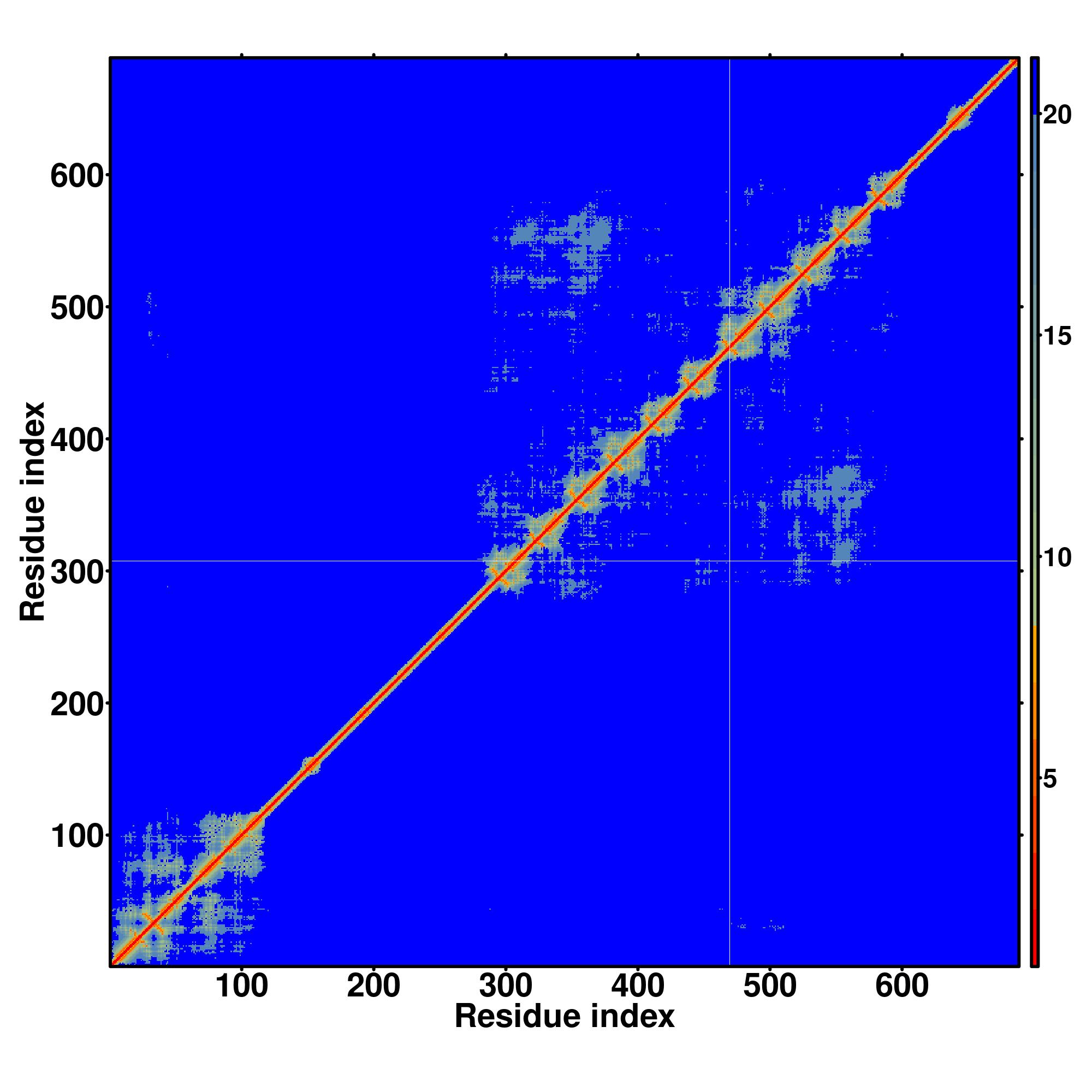
| Rank | PDB
hit | ID1 | ID2 | Cov | Norm.
Zscore | Download
alignment | | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300
| | | | | | | | | | | | | | | |
|---|
| SS
Seq | CSCCCCCCSCCCCCCHHHHCCCCCCCCCCSCCCCCCCSSCCCCCCCCSCCCCCCCCCCSCCCCCCCSCCCCCCCCCSCCCCCCCCCCSCCCCCCCSCCCCCCHHHHCCCSCCCCCSCCCCCCCSCCCCCCHHHHCCCCCCCCCSCCCCCCCSCCCCCCHHHHCCCCCCCCCCSSCCCCCCSCCCCCCHHHHHCCSCCCCCSSCCCCCCSCCCCCCHHHHCCCSCCCCCCCCCCCSCCCCCHHHHCCCSCCCCCSCCCCCCCSSCCCCCCCCCCCCCCCCSCCCCCCCSCCCCCCCCCCCCSCCCCCCCCCC
YKCSSCSQQFMQKKDLQSHMIKLHGAPKPHACPTCAKCFLSRTELQLHEAFKHRGEKLFVCEECGHRASSRNGLQMHIKAKHRNERPHVCEFCSHAFTQKANLNMHLRTHTGEKPFQCHLCGKTFRTQASLDKHNRTHTGERPFSCEFCEQRFTEKGPLLRHVASRHQEGRPHFCQICGKTFKAVEQLRVHVRRHKGVRKFECTECGYKFTRQAHLRRHMEIHDRVENYNPRQRKLRNLIIEDEKMVVVALQPPAELEVGSAEVIVESLAQGGLASQLPGQRLCAEESFTGPGVLEPSLIITAAVPEDCDT |
|---|
| 1 | 3ugmA | 0.04 | 0.04 | 1.78 | 0.83 | DEthreader | | RAALVAKRGGVT--AMEAVHAS--RNALTPLNL--TPAQVLQRLLPVLCHGLTP-QQLPLCQAHGLPPVVAKALETVQRLLPVLCLETVRL-VLVVASHDGGKQGGKQALETVGKQATV-VIASHDGG-QAHDGGAIGGKAGKQALTVIASNGGGKQALTIGGKQALEVQAKLEVRLLPVVASNGGGKQALETVQRLLLETQRLLPVLVASNGGGKQALETVQRLLPVLCQHGLTAQVVAIAS-------V-L-VLCDHGLTLAQV----------------------LCLTQDQVVA------------- |
| 2 | 5v3jE | 0.30 | 0.26 | 7.89 | 6.03 | SPARKS-K | | HKCKECGKAFHTPSQLSHHQKLHVGE-KPYKCQECGKAFPSNAQLSLHHRVHT-DEKCFECKECGKAFMRPSHLLRHQRIHTG-EKPHKCKECGKAFRYDTQLSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQS-VHTGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELFRHQKVHTGDRPHKCKELTHHERSHSGEK----PYECKECGKTFGRGSELSRHQKIHT------------------------------------- |
| 3 | 5v3jE | 0.31 | 0.27 | 8.18 | 1.42 | MapAlign | | HKCKECGKAFHTPSQLSHHQKLHV-GEKPYKCQECGKAFPSNAQLSLHH-RVHTDEKCFECKECGKAFMRPSHLLRHQRI-HTGEKPHKCKECGKAFRYDTQLSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVH-TGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELFRHQKVHTGD----------RPHKCKECGKAFIR------------RSELTHHE-RSHSGEKPYECKECGKTFGRGSELSRHQKIH--------- |
| 4 | 5v3jE | 0.31 | 0.24 | 7.40 | 1.05 | CEthreader | | YKCQECGKAFPSNAQLSLHHRVHT-DEKCFECKECGKAFMRPSHLLRHQRI-HTGEKPHKCKECGKAFRYDTQLSLHLL-THAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHQR-AHSGDKPYKCKECGKSFTCTTELFRHQKVHTGDRPHKCKECGKAFIRRSELTHHERSHSGEKPYECKECGKTFGRGSELSRHQKIHT----------------------------------------------------------- |
| 5 | 5v3jE | 0.31 | 0.27 | 8.16 | 3.54 | MUSTER | | HKCKECGKAFHTPSQLSHHQKLH-VGEKPYKCQECGKAFPSNAQLSLHHRV-HTDEKCFECKECGKAFMRPSHLLRHQRIH-TGEKPHKCKECGKAFRYDTQLSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVH-TGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELFRHQKVHTGDRPHKCKECFIRRSELTHHERSHSGEKPYECKECGKTFGRGSELSQKIHT------------------------------------ |
| 6 | 5v3jE | 0.33 | 0.25 | 7.54 | 2.24 | HHsearch | | YKCQECGKAFPSNAQLSLHH-RVHTDEKCFECKECGKAFMRPSHLLRHQRI-HTGEKPHKCKECGKAFRYDTQLSLHLLT-HAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHQR-AHSGDKPYKCKECGKSFTCTTELFRHQKVHTGDRPHKCKECGKAFIRRSELTHHERSHSGEKPYECKECELSRHQK------IHT------------------------------------------------------------- |
| 7 | 5v3jE | 0.32 | 0.28 | 8.34 | 2.72 | FFAS-3D | | HKCKECGKAFHTPSQLSHHQ-KLHVGEKPYKCQECGKAFPSNAQLSLHHRV-HTDEKCFECKECGKAFMRPSHLLRHQRIH-TGEKPHKCKECGKAFRYDTQLSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVH-TGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELFRHQKVHTGDRPHKCKECFIRRSELTHHERSHSGEKPYECKECGKTFGRGSELSRHQKIHT---------------------------------- |
| 8 | 5v3jE | 0.35 | 0.24 | 7.31 | 1.27 | EigenThreader | | HKCKECGKAFHTPSQLSHHQKLHVGE-KPYKCQECGKAFPSNAQLSLHHRVHTDEKCFECK-ECGKAFMRPSHLLRHQRIHTG-EKPHKCKECGKAFRYDTQLSLHLLTHAGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELFRHQKVHTGDRPHKCKEC-GKAFIRRSELTHHERSHSGEKPYECKECGKTFGRGSELSRHQKIHT--------------------------------------------------------------------------------------- |
| 9 | 5v3mC | 0.33 | 0.24 | 7.33 | 6.72 | CNFpred | | YKCQECGKAFPSNAQLSLHHRVHTD-EKCFECKECGKAFMRPSHLLRHQR-IHTGEKPHKCKECGKAFRYDTQLSLHLL-THAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHQR-AHSGDKPYKCKECGKSFTCTTELFRHQKVHTGDRPHKCKECGKAFIRRSELTHHERSHSGEKPYECK------------------------------------------------------------------------------- |
| 10 | 5v3jE | 0.29 | 0.19 | 5.88 | 0.83 | DEthreader | | ---------------------RVHTDEKCFECKECGKAFMRPSHLLRHQ-RIHT-GEKPHKCECGKAFRY-DTQLSLHLLTHAGARRFECK-DCDKVYSASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELA-RHQRAHSGDKPYKCKECGKSFTCTTELFRHQKVHTGDRPHKCKECGKAFIRRSELTHHERSH-SG-EK---P---Y--E---------CGKT--------------------------------IH------------------------ |
| (a) | ID1 is the number of template residues identical to query divided by number of aligned residues. |
| (b) | ID2 is the number of template residues identical to query divided by query sequence length. |
| (c) | Cov is equal the number of aligned template residues divided by query sequence length. |
| (d) | Norm. Zscore is the normalized Z-score of the threading alignments. A Normalized Z-score >1 means a good alignment and is highlighted in bold. |
| (e) | Download alignment lists the threading program used to identify the template, and provide the 3D structure of aligned regions of threading templates (threading[1-10].pdb.gz). |
| (f) | Template residues identical to query sequence are highlighted in color. |
|