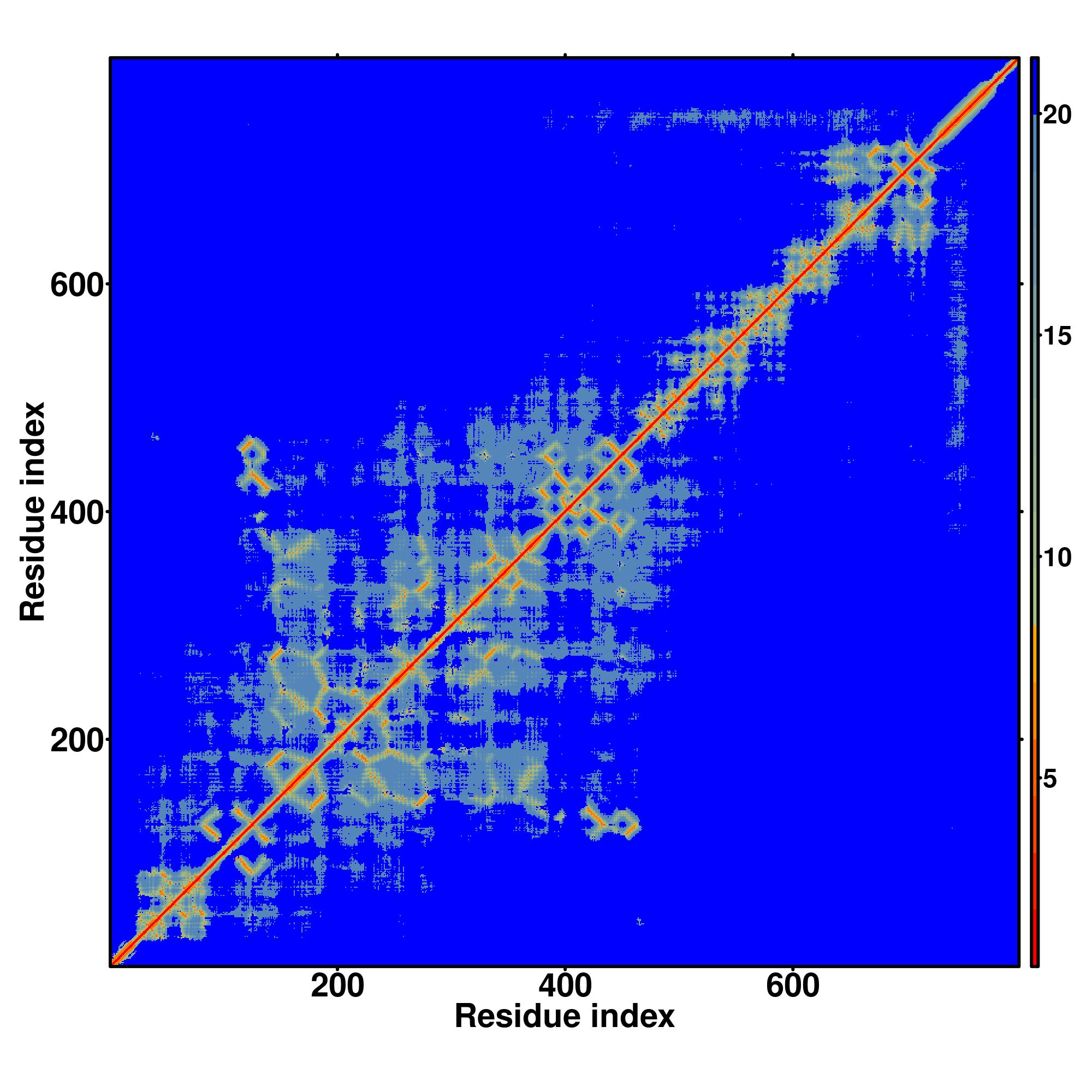
| GO term | CscoreGO | Name |
| GO:0044699 | 0.97 | single-organism process |
| GO:0044763 | 0.95 | single-organism cellular process |
| GO:0065007 | 0.83 | biological regulation |
| GO:0050789 | 0.83 | regulation of biological process |
| GO:0050794 | 0.81 | regulation of cellular process |
| GO:0050896 | 0.65 | response to stimulus |
| GO:0032879 | 0.65 | regulation of localization |
| GO:0016043 | 0.65 | cellular component organization |
| GO:1902578 | 0.63 | single-organism localization |
| GO:0048518 | 0.63 | positive regulation of biological process |
| GO:0060255 | 0.61 | regulation of macromolecule metabolic process |
| GO:0071495 | 0.60 | cellular response to endogenous stimulus |
| GO:0071310 | 0.60 | cellular response to organic substance |
| GO:0071704 | 0.59 | organic substance metabolic process |
| GO:0048522 | 0.59 | positive regulation of cellular process |
| GO:0044765 | 0.59 | single-organism transport |
| GO:0044710 | 0.59 | single-organism metabolic process |
| GO:0016192 | 0.59 | vesicle-mediated transport |
| GO:0071402 | 0.58 | cellular response to lipoprotein particle stimulus |
| GO:0055098 | 0.58 | response to low-density lipoprotein particle |
| GO:0043170 | 0.58 | macromolecule metabolic process |
| GO:0006898 | 0.58 | receptor-mediated endocytosis |
| GO:0051128 | 0.56 | regulation of cellular component organization |
| GO:0051239 | 0.54 | regulation of multicellular organismal process |
| GO:0007155 | 0.54 | cell adhesion |
| GO:0051130 | 0.53 | positive regulation of cellular component organization |
| GO:0044237 | 0.52 | cellular metabolic process |
| GO:0007229 | 0.52 | integrin-mediated signaling pathway |
| GO:0071404 | 0.51 | cellular response to low-density lipoprotein particle stimulus |
| GO:0031623 | 0.51 | receptor internalization |