|
| GO term | CscoreGO | Name |
| GO:1901363 | 0.87 | heterocyclic compound binding |
| GO:0097159 | 0.87 | organic cyclic compound binding |
| GO:0003677 | 0.86 | DNA binding |
| GO:0043565 | 0.85 | sequence-specific DNA binding |
| GO:0046982 | 0.84 | protein heterodimerization activity |
| GO:0042802 | 0.84 | identical protein binding |
| GO:0000989 | 0.84 | transcription factor activity, transcription factor binding |
| GO:0000976 | 0.84 | transcription regulatory region sequence-specific DNA binding |
| GO:0042803 | 0.83 | protein homodimerization activity |
| GO:0001106 | 0.83 | RNA polymerase II transcription corepressor activity |
| GO:0003700 | 0.58 | transcription factor activity, sequence-specific DNA binding |
| GO:0000981 | 0.53 | RNA polymerase II transcription factor activity, sequence-specific DNA binding |
| GO:0000977 | 0.52 | RNA polymerase II regulatory region sequence-specific DNA binding |
| GO:0001078 | 0.50 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding |
| GO:0000978 | 0.50 | RNA polymerase II core promoter proximal region sequence-specific DNA binding |
| Download full result of the above consensus prediction. |
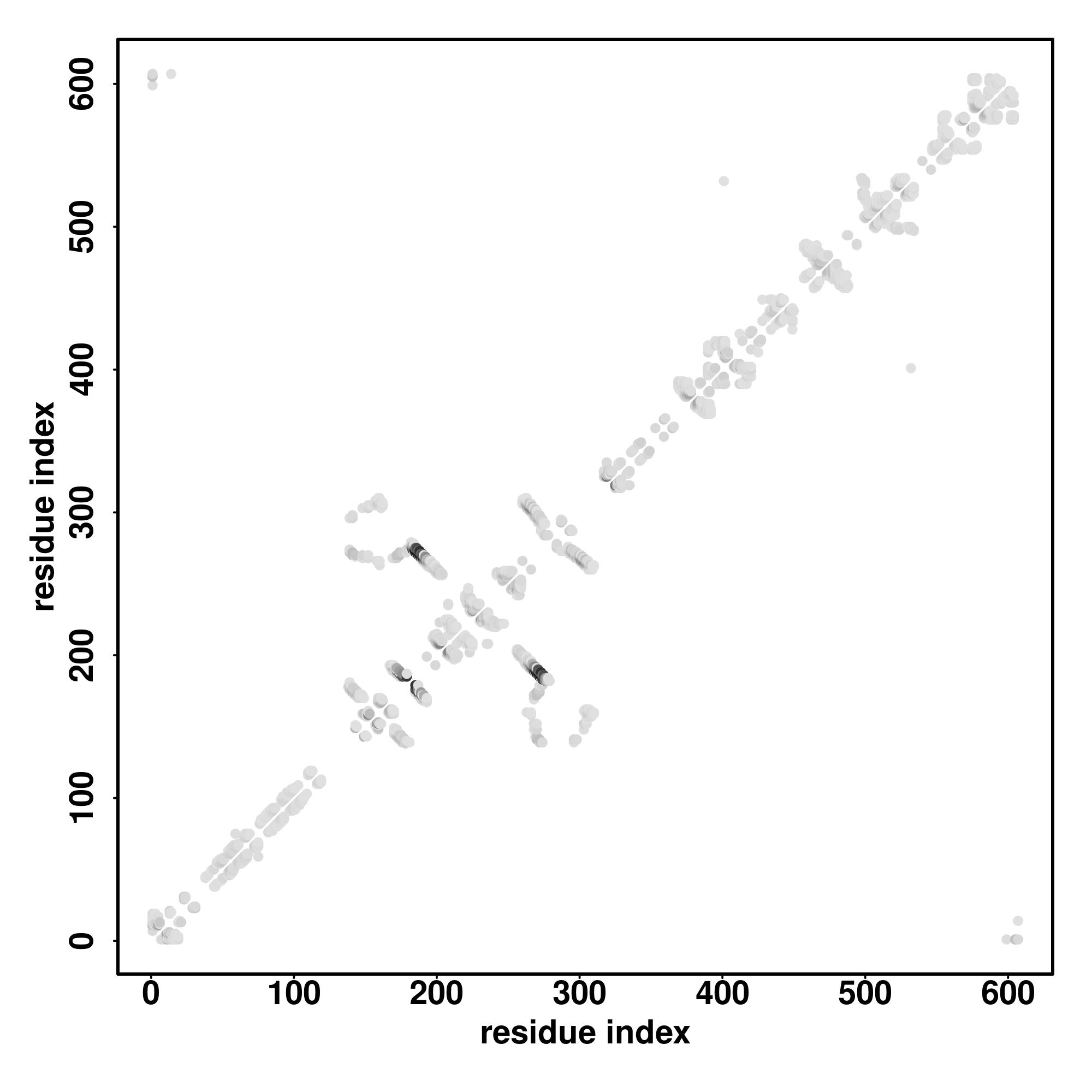
| Click the graph to show a high resolution version. |
| (a) | CscoreGO is the confidence score of predicted GO terms. CscoreGO values range in between [0-1]; where a higher value indicates a better confidence in predicting the function using the template. |
| (b) | The graph shows the predicted terms within the Gene Ontology hierachy for Molecular Function. Confidently predicted terms are color coded by CscoreGO: |
| | [0.4,0.5) | [0.5,0.6) | [0.6,0.7) | [0.7,0.8) | [0.8,0.9) | [0.9,1.0] |
|
|

|
| GO term | CscoreGO | Name |
| GO:0032502 | 1.00 | developmental process |
| GO:0044699 | 0.82 | single-organism process |
| GO:0065007 | 0.76 | biological regulation |
| GO:0044767 | 0.76 | single-organism developmental process |
| GO:0050789 | 0.75 | regulation of biological process |
| GO:0050794 | 0.73 | regulation of cellular process |
| GO:0048856 | 0.67 | anatomical structure development |
| GO:0051239 | 0.65 | regulation of multicellular organismal process |
| GO:0050793 | 0.65 | regulation of developmental process |
| GO:0048583 | 0.65 | regulation of response to stimulus |
| GO:0048523 | 0.65 | negative regulation of cellular process |
| GO:0023051 | 0.65 | regulation of signaling |
| GO:2000026 | 0.64 | regulation of multicellular organismal development |
| GO:0022603 | 0.64 | regulation of anatomical structure morphogenesis |
| GO:0009968 | 0.64 | negative regulation of signal transduction |
| GO:0072001 | 0.63 | renal system development |
| GO:0060829 | 0.63 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation |
| GO:0048646 | 0.55 | anatomical structure formation involved in morphogenesis |
| GO:0009653 | 0.53 | anatomical structure morphogenesis |
| Download full result of the above consensus prediction. |
| Click the graph to show a high resolution version. |
| (a) | CscoreGO is the confidence score of predicted GO terms. CscoreGO values range in between [0-1]; where a higher value indicates a better confidence in predicting the function using the template. |
| (b) | The graph shows the predicted terms within the Gene Ontology hierachy for Biological Process. Confidently predicted terms are color coded by CscoreGO: |
| | [0.4,0.5) | [0.5,0.6) | [0.6,0.7) | [0.7,0.8) | [0.8,0.9) | [0.9,1.0] |
|
|

|
| Download full result of the above consensus prediction. |
| Click the graph to show a high resolution version. |
| (a) | CscoreGO is the confidence score of predicted GO terms. CscoreGO values range in between [0-1]; where a higher value indicates a better confidence in predicting the function using the template. |
| (b) | The graph shows the predicted terms within the Gene Ontology hierachy for Cellular Component. Confidently predicted terms are color coded by CscoreGO: |
| | [0.4,0.5) | [0.5,0.6) | [0.6,0.7) | [0.7,0.8) | [0.8,0.9) | [0.9,1.0] |
|
|
|