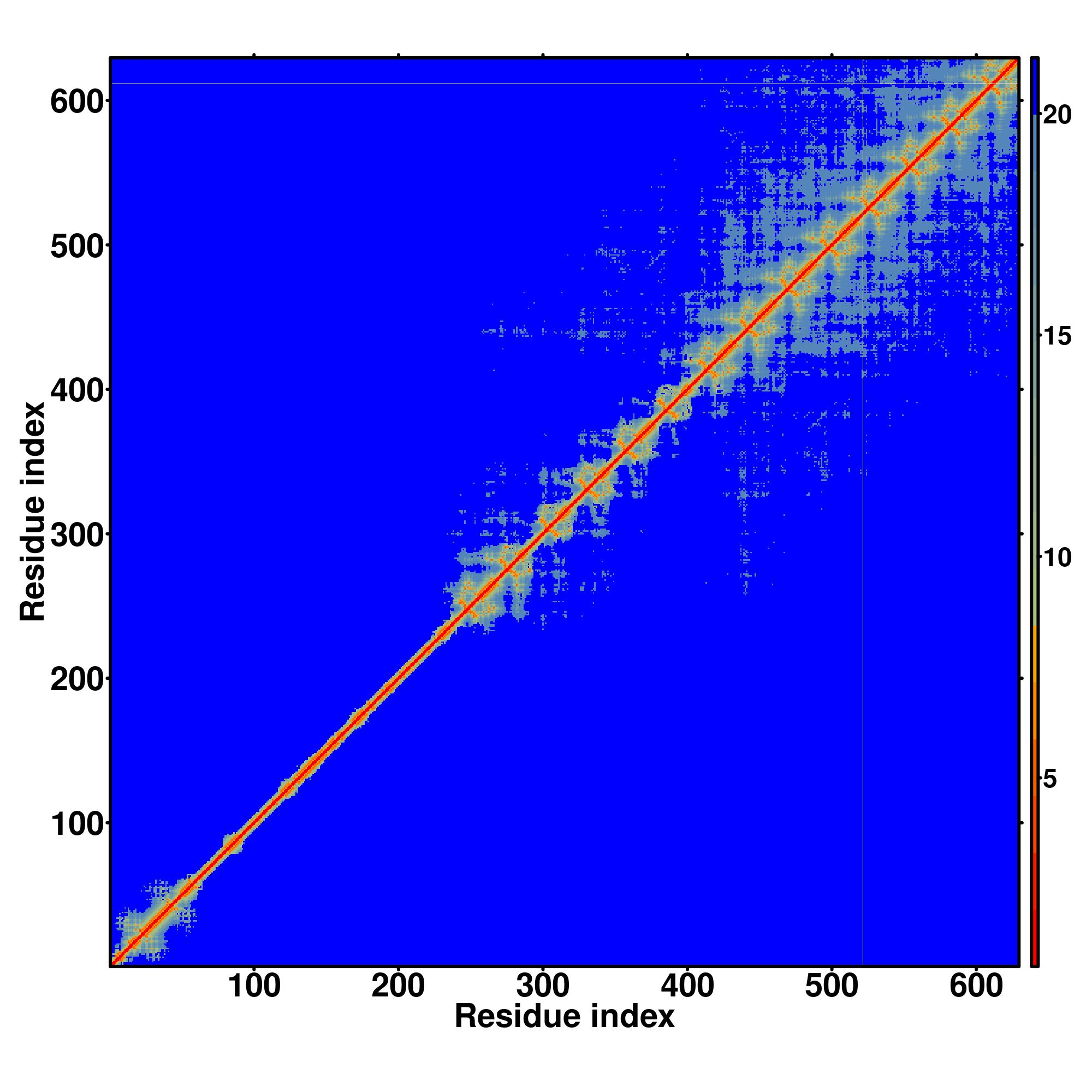
| Rank | PDB
hit | ID1 | ID2 | Cov | Norm.
Zscore | Download
alignment | | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300
| | | | | | | | | | | | | | | |
|---|
| SS
Seq | CCCCCCCCCCCCCSCSCCCCCCCCCCHHHHCCCCCHHHCCCCCCHHHHHHHCHHCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSCCCCCCCCCSCCCCCSSCCCCCCSCCCCCCCSSCCCCCCCCSSCCCCCCSSCCCCCCHHHCCCSCCCCCSCCCCCCCSSCCCCCHHHHCCCSCCCCCSCCCCCCCSSCCCCCCCCSCCCCCCCCCSCCCCCCCSSCCCCCCCCCCCSCCCCCSCCCCCCCSSCCCCCCCCCCCCCCCCCSCCCCCCCSSCCCCCHHHHHHCCCCCCC
PEMGFHHATQACLELLGSSDLPASASQSAGITGVNHRAQPGLNVSVDKFTALCSPGVLQTVKWFLEFRCIFSLAMSSHFTQDLLPEQGIQDAFPKRILRGYGNCGLDNLYLRKDWESLDECKLQKDYNGLNQCSSTTHSKIFQYNKYVKIFDNFSNLHRRNISNTGEKPFKCQECGKSFQMLSFLTEHQKIHTGKKFQKCGECGKTFIQCSHFTEPENIDTGEKPYKCQECNNVIKTCSVLTKNRIYAGGEHYRCEEFGKVFNQCSHLTEHEHGTEEKPCKYEECSSVFISCSSLSNQQMILAGEKL |
|---|
| 1 | 5mzsA | 0.05 | 0.04 | 1.71 | 0.67 | DEthreader | | APPSNDLSQII-QILVPVSSRNGRSRGDTN-VAVRIEVIRGPAAAGGVV-NIIT-Q--RVYGNDIDGFSRQ-GN--IYT-GDTQTNSNNYVKQML-G-------HETN------------INEGLAG-GTEGIFDNAGFYTATLRDLKLDDPSSN-SSSSSARIFHHDIVGDNWSPYKAPNQLLKAETSDYKNPKAVVEGLTYMLQSKNVLSVTPRYT----LNSML---QATVTWYGQ-PYAIADNLFDKR----------LF--RAGNAQG--VVGID----------GAG-------------- |
| 2 | 5v3jE | 0.29 | 0.22 | 6.75 | 5.49 | SPARKS-K | | ------------------------------------------------------------------------PHKCKECGKAFHTPSQLSHHQKLEKPYKCQECGKSNAQLSLHHRVHTDEKCFEKECGLRHQRIHTGEKPHKCKECGKAFRYDTQLSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHRAHSGDKPYKCKECGKSFTCTTELFRHQKVTGDRPHKCKECGKAFIRRSELTHHERSHSGEKP |
| 3 | 5v3jE | 0.27 | 0.24 | 7.29 | 1.29 | MapAlign | | --------------------------PHKCKECGKAFHTPSQLSHHQKLHVGEKPYKCQECGKAFPSNAQLSLHHRVHTDEKCFECKECGKAF----------MRPSHLLRHQRIHTGEKPHKCKECGKAFHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELFRHQKVHTDRPHKCKECGKAFIRRSELTHHERSHSEKPYECKECGKTFGRGSELSRHQKIH----- |
| 4 | 5v3jE | 0.29 | 0.22 | 6.85 | 0.84 | CEthreader | | ------------------------------------------------------------------PHKCKECGKAFHTPSQLSHHQKLHVGEKPYKCQECGKAFPSNAQLSLHHRVHTDEKCFRPSHLLRHQRIHTGEKPHKCKECGKAFRYDTQLSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHRAHSGDKPYKCKECGKSFTCTTELFRHQKHTGDRPHKCKECGKAFIRRSELTHHERSHSGEKP |
| 5 | 5v3jE | 0.26 | 0.23 | 7.03 | 3.59 | MUSTER | | -----------------------------------PHKCKECGKAFHTPSQLSHHKLHVGEKPYKCQECGKAFPSNAQLSLHHRVHTDEKCFECKECGKAFMRPSHQRIHTGEKPHKCKECGKAFRYDTSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELFRHKVHTGDRPHKCKECGKAFIRRSELTHHERHSGEKPYECKECGKTFGRGSELSRHQKIHT---- |
| 6 | 5v3jE | 0.30 | 0.24 | 7.42 | 1.94 | HHsearch | | -----------------------------------PHKCKEC------GKAFHTPSQLSEKPYK----CQECGK---AFPSAQLSLH--HRVHTDEKCFECKECGKRPSHLLRHQRIHTGEKPHKKECGSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELFRHQVHTGDRPHKCKECGKAFIRRSELTHHERHSGEKPYECKECGKTFGRGSELSRHQKIHT---- |
| 7 | 5v3jE | 0.26 | 0.22 | 6.94 | 2.28 | FFAS-3D | | ----------------------------------PHKCKECGKAFHTPSQLSHHQKLHVGEKPYKCQECGKAFPSNAQLSLHHRVHTDEKCFECKECGKAFMRPSHQRIHTGEKPHKCKECGKAFRTQLSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELRHQKVHTGDRPHKCKECGKAFIRRSELTHHERHSGEKPYECKECGKTFGRGSELSRHQKIHT---- |
| 8 | 5v3jE | 0.22 | 0.17 | 5.45 | 1.02 | EigenThreader | | GKAFHTPSQLSHHQKLHVGEKPYKCQECGKAFPS------NAQLSLHHRVHTDEKCFECKECGK-------------AFMRPSHLLRHQRIHTGEKPHKCKECGKAYDTQLSLHLLT-HAGARRFECKDCDKVYSCTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHQRAHSGDKPYKCKECGKSFTCTTELFRHQKVHTGDRPHKCKECGKAFIRRSELTHHERSHSGEKCKEC--GKTFGRT------------------------------------------ |
| 9 | 5v3mC | 0.31 | 0.21 | 6.39 | 7.87 | CNFpred | | --------------------------------------------------------------------------------------------------YKCQECGKAFPQLSLHHRVHTDEKCFECECGKA-QRIHTGEKPHKCKECGKAFRYDTQLSLHLLTHAGARRFECKDCDKVYSCASQLALHQMSHTGEKPHKCKECGKGFISDSHLLRHQSVHTGETPYKCKECGKGFRRGSELARHQAHSGDKPYKCKECGKSFTCTTELFRHQKHTGDRPHKCKECGKAFIRRSELTHHERSHSGEKP |
| 10 | 4cr2Z | 0.06 | 0.04 | 1.84 | 0.67 | DEthreader | | ----------------------------------TD-IALAVRLG-EAQKTSFEEQLTGPKVPEDIYK---------F-SAGLDSAQQNLASSF-VNGFLNLGY-CNDKLIVDND--NWVYKTKGDG--MTSAVAVKAGALLIG--VEPALLAAILGLGFDEVLGLLLPAAS--CNGDITSIWFLAALGVLTISTSAVLSVLIQDLRYAVLGIALIALGEDIGKEMS--------IVSVSPCGATNNARLQLLRASYYS----ALFITRLA-------------------GLVSPSFMLK------- |
| (a) | ID1 is the number of template residues identical to query divided by number of aligned residues. |
| (b) | ID2 is the number of template residues identical to query divided by query sequence length. |
| (c) | Cov is equal the number of aligned template residues divided by query sequence length. |
| (d) | Norm. Zscore is the normalized Z-score of the threading alignments. A Normalized Z-score >1 means a good alignment and is highlighted in bold. |
| (e) | Download alignment lists the threading program used to identify the template, and provide the 3D structure of aligned regions of threading templates (threading[1-10].pdb.gz). |
| (f) | Template residues identical to query sequence are highlighted in color. |
|