

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MACNSTSLEAYTYLLLNTSNASDSGSTQLPAPLRISLAIVMLLMTVVGFLGNTVVCIIVYQRPAMRSAINLLLATLAFSDIMLSLCCMPFTAVTLITVRWHFGDHFCRLSATLYWFFVLEGVAILLIISVDRFLIIVQRQDKLNPRRAKVIIAVSWVLSFCIAGPSLTGWTLVEVPARAPQCVLGYTELPADRAYVVTLVVAVFFAPFGVMLCAYMCILNTVRKNAVRVHNQSDSLDLRQLTRAGLRRLQRQQQVSVDLSFKTKAFTTILILFVGFSLCWLPHSVYSLLSVFSQRFYCGSSFYATSTCVLWLSYLKSVFNPIVYCWRIKKFREACIELLPQTFQILPKVPERIRRRIQPSTVYVCNENQSAV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 998889875444566787888888888888799999999999999999998998623305878998599999999999999999999999999999585518448987999999999999999999999973515674898887778989446999999999999999732001479976451678885314179999999999999999999999999999997620124322100110023444443124566689998889999999999999999899999999999817446873289999999999999999989999838999999999985422467888987556777898874568899999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MACNSTSLEAYTYLLLNTSNASDSGSTQLPAPLRISLAIVMLLMTVVGFLGNTVVCIIVYQRPAMRSAINLLLATLAFSDIMLSLCCMPFTAVTLITVRWHFGDHFCRLSATLYWFFVLEGVAILLIISVDRFLIIVQRQDKLNPRRAKVIIAVSWVLSFCIAGPSLTGWTLVEVPARAPQCVLGYTELPADRAYVVTLVVAVFFAPFGVMLCAYMCILNTVRKNAVRVHNQSDSLDLRQLTRAGLRRLQRQQQVSVDLSFKTKAFTTILILFVGFSLCWLPHSVYSLLSVFSQRFYCGSSFYATSTCVLWLSYLKSVFNPIVYCWRIKKFREACIELLPQTFQILPKVPERIRRRIQPSTVYVCNENQSAV | |
| 642120213323312233122343433312200100001111210231333321000000003301100100020003021120100000100200264300130011000000000000003030000010000002024302330000000000110001001000002223344422201130123212200000002211320010002001100010233034144344434444444444443444443333133320000000000000000311110000000003302243001001000001002000310001000164004001400200231145345434343444333314544334 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MACNSTSLEAYTYLLLNTSNASDSGSTQLPAPLRISLAIVMLLMTVVGFLGNTVVCIIVYQRPAMRSAINLLLATLAFSDIMLSLCCMPFTAVTLITVRWHFGDHFCRLSATLYWFFVLEGVAILLIISVDRFLIIVQRQDKLNPRRAKVIIAVSWVLSFCIAGPSLTGWTLVEVPARAPQCVLGYTELPADRAYVVTLVVAVFFAPFGVMLCAYMCILNTVRKNAVRVHNQSDSLDLRQLTRAGLRRLQRQQQVSVDLSFKTKAFTTILILFVGFSLCWLPHSVYSLLSVFSQRFYCGSSFYATSTCVLWLSYLKSVFNPIVYCWRIKKFREACIELLPQTFQILPKVPERIRRRIQPSTVYVCNENQSAV | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.19 | 0.21 | 0.77 | 2.60 | Download | ---------------ENFMDIECF--MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNC---EKLQSVCSDIFPHID---ETYLMFWIGVTSVLLLFIVYAYMYILWKAG--------------------------KRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGK---MNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------------------------- | |||||||||||||||||||
| 2 | 5uenA | 0.21 | 0.23 | 0.83 | 4.09 | Download | -------------------------------AFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYFH--TCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLSGSMGEPVIKCEFEKVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDNVKDARNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCHKP---SILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRC--------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.19 | 0.21 | 0.80 | 3.23 | Download | ---------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------------------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.19 | 0.22 | 0.95 | 1.53 | Download | ---DLRDNETWWYNPSIIVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAAMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNA-----------KELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV---TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQDKETEDAETEIPSSDAAPSADQMKE | |||||||||||||||||||
| 5 | 3uon | 0.21 | 0.24 | 0.81 | 1.18 | Download | ------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVVEDGECYIQFF---SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGAAIGRNTNVNAAGNRWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC----IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.19 | 0.21 | 0.79 | 2.88 | Download | -----------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV------------------------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.21 | 0.22 | 0.82 | 1.71 | Download | ------------------------EFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAAMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNA-----------KELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVT---PYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWV------------------------------ | |||||||||||||||||||
| 8 | 1u19A | 0.20 | 0.24 | 0.90 | 4.58 | Download | M--NGTEGPNFYVPFSNKTGVVRSPFEAPEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETSFVIYMFVVHFIIPLIVIFFCYG-----------------------QLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIP---AFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCC---------GKNPLGDDEASTTVSKTETSQV | |||||||||||||||||||
| 9 | 3sn6R | 0.24 | 0.20 | 0.76 | 3.03 | Download | ------------------------------EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKLLTKNKARVIILMVWIVSGLTSFLPIQM-HWYRQEAINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR-----------------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIR----KEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC-------------------------------- | |||||||||||||||||||
| 10 | 3zpqA | 0.26 | 0.24 | 0.76 | 3.61 | Download | ------------------------GAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQ-----------------------------SRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP----DWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLLA-------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
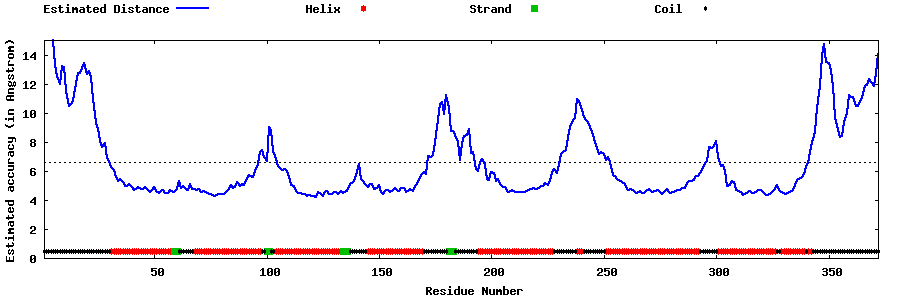
| Generated 3D models | Estimated local accuracy of models | ||
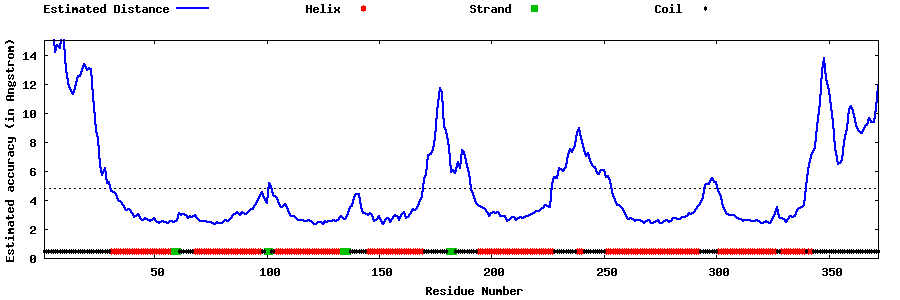 | |||
|
|
|||
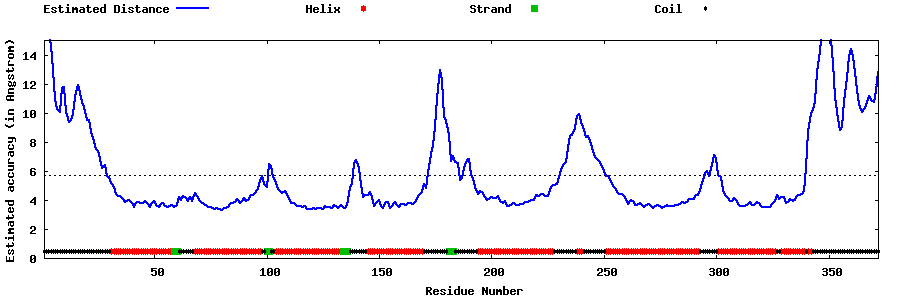 | |||
|
| |||
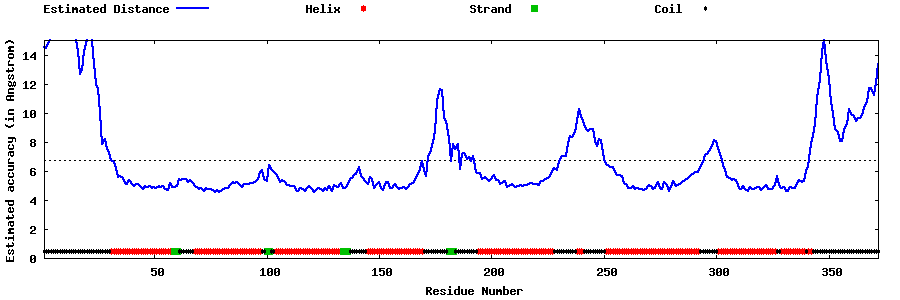 | |||
|
| |||
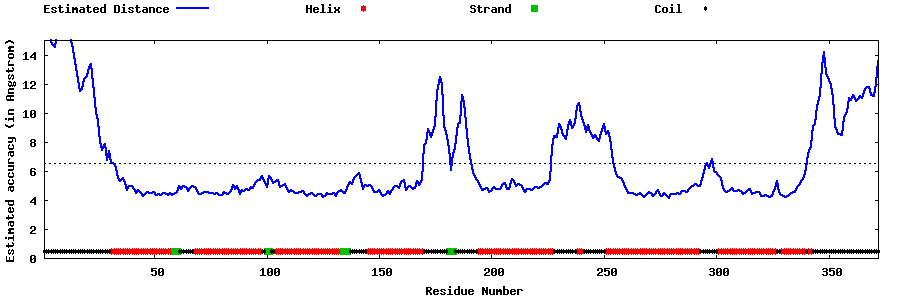 | |||
|
| |||
 | |||
|
| |||