

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDETGNLTVSSATCHDTIDDFRNQVYSTLYSMISVVGFFGNGFVLYVLIKTYHKKSAFQVYMINLAVADLLCVCTLPLRVVYYVHKGIWLFGDFLCRLSTYALYVNLYCSIFFMTAMSFFRCIAIVFPVQNINLVTQKKARFVCVGIWIFVILTSSPFLMAKPQKDEKNNTKCFEPPQDNQTKNHVLVLHYVSLFVGFIIPFVIIIVCYTMIILTLLKKSMKKNLSSHKKAIGMIMVVTAAFLVSFMPYHIQRTIHLHFLHNETKPCDSVLRMQKSVVITLSLAASNCCFDPLLYFFSGGNFRKRLSTFRKHSLSSVTYVPRKKASLPEKGEEICKV | |
| CCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9998899999877776447788987999999999999988999778776246768699999999999999999978999999956998887856899999999999999999999999971277740030124656204765679999999999989999301463799658816699521279999999999999999999999999999999983366666641011289999999999999856899999999999836789917999999999999999999998989999940888999999999875267877888788999998755379 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDETGNLTVSSATCHDTIDDFRNQVYSTLYSMISVVGFFGNGFVLYVLIKTYHKKSAFQVYMINLAVADLLCVCTLPLRVVYYVHKGIWLFGDFLCRLSTYALYVNLYCSIFFMTAMSFFRCIAIVFPVQNINLVTQKKARFVCVGIWIFVILTSSPFLMAKPQKDEKNNTKCFEPPQDNQTKNHVLVLHYVSLFVGFIIPFVIIIVCYTMIILTLLKKSMKKNLSSHKKAIGMIMVVTAAFLVSFMPYHIQRTIHLHFLHNETKPCDSVLRMQKSVVITLSLAASNCCFDPLLYFFSGGNFRKRLSTFRKHSLSSVTYVPRKKASLPEKGEEICKV | |
| 6254443434443144324301110002201200231332322001000122342200000013311030000000001000003544210010011001111131111101000000310010000002024223221000000001100000010100012035572211001113673141010011122123013312210030001001101325456445422000000000000012013311210001002313324414133002101100100013000300100000045014201400342244354354454444544433367 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC MDETGNLTVSSATCHDTIDDFRNQVYSTLYSMISVVGFFGNGFVLYVLIKTYHKKSAFQVYMINLAVADLLCVCTLPLRVVYYVHKGIWLFGDFLCRLSTYALYVNLYCSIFFMTAMSFFRCIAIVFPVQNINLVTQKKARFVCVGIWIFVILTSSPFLMAKPQKDEKNNTKCFEPPQDNQTKNHVLVLHYVSLFVGFIIPFVIIIVCYTMIILTLLKKSMKKNLSSHKKAIGMIMVVTAAFLVSFMPYHIQRTIHLHFLHNETKPCDSVLRMQKSVVITLSLAASNCCFDPLLYFFSGGNFRKRLSTFRKHSLSSVTYVPRKKASLPEKGEEICKV | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.23 | 0.24 | 0.88 | 3.40 | Download | -----------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK--EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 2 | 4xnwA | 0.31 | 0.30 | 0.88 | 4.48 | Download | ---------SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTVRKNKTITCYDTTSDEY-LRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLQTPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR----------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.24 | 0.24 | 0.88 | 3.97 | Download | -----------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRM--KEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.29 | 0.29 | 0.84 | 1.54 | Download | ------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEA--LGS---A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------- | |||||||||||||||||||
| 5 | 5t1a | 0.26 | 0.25 | 0.85 | 1.16 | Download | -----------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKE-DSVYVCGPYFPRG----WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIPPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF--------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.23 | 0.24 | 0.88 | 3.52 | Download | ------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE--KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.29 | 0.29 | 0.82 | 1.70 | Download | ---------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF-------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.24 | 0.24 | 0.87 | 4.75 | Download | -----------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREKKRHRD------VRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.23 | 0.24 | 0.88 | 3.53 | Download | -----------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE--KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.25 | 0.27 | 0.91 | 3.97 | Download | RNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSP-SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
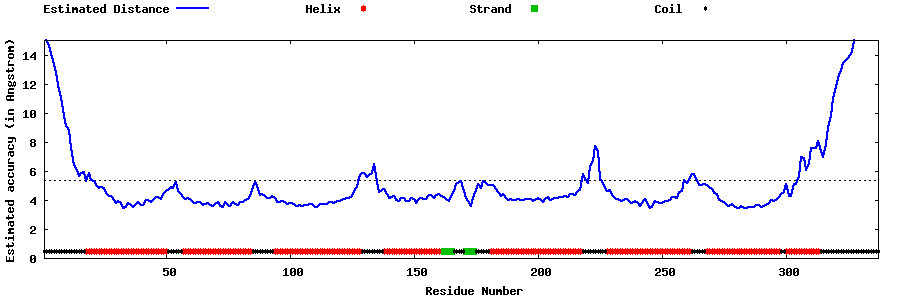
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||