

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MCPMLLKNGYNGNATPVTTTAPWASLGLSAKTCNNVSFEESRIVLVVVYSAVCTLGVPANCLTAWLALLQVLQGNVLAVYLLCLALCELLYTGTLPLWVIYIRNQHRWTLGLLACKVTAYIFFCNIYVSILFLCCISCDRFVAVVYALESRGRRRRRTAILISACIFILVGIVHYPVFQTEDKETCFDMLQMDSRIAGYYYARFTVGFAIPLSIIAFTNHRIFRSIKQSMGLSAAQKAKVKHSAIAVVVIFLVCFAPYHLVLLVKAAAFSYYRGDRNAMCGLEERLYTASVVFLCLSTVNGVADPIIYVLATDHSRQEVSRIHKGWKEWSMKTDVTRLTHSRDTEELQSPVALADHYTFSRPVHPPGSPCPAKRLIEESC | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSSSSHHHHHHHHHHHHHHHSSSCCCSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99866688888989977777888888888765666554147888999999999999988988313010326778699999999999999999986999999955899888865699999999999999999999999970044120303214667633231149999999999746579889527707799147899999999999999999999999999999983567877542016899999999999998678999999999999740356777725899999999999999999998988988714889999999998642335556788878888987655787656788778888899999998565882419 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MCPMLLKNGYNGNATPVTTTAPWASLGLSAKTCNNVSFEESRIVLVVVYSAVCTLGVPANCLTAWLALLQVLQGNVLAVYLLCLALCELLYTGTLPLWVIYIRNQHRWTLGLLACKVTAYIFFCNIYVSILFLCCISCDRFVAVVYALESRGRRRRRTAILISACIFILVGIVHYPVFQTEDKETCFDMLQMDSRIAGYYYARFTVGFAIPLSIIAFTNHRIFRSIKQSMGLSAAQKAKVKHSAIAVVVIFLVCFAPYHLVLLVKAAAFSYYRGDRNAMCGLEERLYTASVVFLCLSTVNGVADPIIYVLATDHSRQEVSRIHKGWKEWSMKTDVTRLTHSRDTEELQSPVALADHYTFSRPVHPPGSPCPAKRLIEESC | |
| 71313233423343333333333343424443043343501100002001200230231011000000122343210000000000000000000101000002433210121011001211121010000000000200000001002024223321000000000100000000101044322021303372020001121113012211200020000001101314546543211000000000000000122101000010023022333333304123002001100100012000110201000034015101400342343346464443444543543444443454433445443674423365246736 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSSSSHHHHHHHHHHHHHHHSSSCCCSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MCPMLLKNGYNGNATPVTTTAPWASLGLSAKTCNNVSFEESRIVLVVVYSAVCTLGVPANCLTAWLALLQVLQGNVLAVYLLCLALCELLYTGTLPLWVIYIRNQHRWTLGLLACKVTAYIFFCNIYVSILFLCCISCDRFVAVVYALESRGRRRRRTAILISACIFILVGIVHYPVFQTEDKETCFDMLQMDSRIAGYYYARFTVGFAIPLSIIAFTNHRIFRSIKQSMGLSAAQKAKVKHSAIAVVVIFLVCFAPYHLVLLVKAAAFSYYRGDRNAMCGLEERLYTASVVFLCLSTVNGVADPIIYVLATDHSRQEVSRIHKGWKEWSMKTDVTRLTHSRDTEELQSPVALADHYTFSRPVHPPGSPCPAKRLIEESC | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.19 | 0.17 | 0.76 | 3.05 | Download | ------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTLHYTCSSHFPYSFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE-KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQ----EFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------------------------------------- | |||||||||||||||||||
| 2 | 4xnwA | 0.30 | 0.24 | 0.77 | 3.79 | Download | ----------------------------SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYNKTITCYDTTSDEYSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFQ--TPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR----------------------------------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.20 | 0.17 | 0.76 | 3.46 | Download | ------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ--WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE-KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN----CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.17 | 0.19 | 0.74 | 1.51 | Download | -----------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGENITCVLTKER---FGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADTISNEQRASKVLGIVFFLFLLMWCPFFITNITLV--LCDSC--------NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---------------------------------------------------- | |||||||||||||||||||
| 5 | 5t1a | 0.21 | 0.21 | 0.74 | 1.16 | Download | ------------------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTSVYVCGPYFPRG-WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFF----GLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF--------------------------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.19 | 0.17 | 0.75 | 3.09 | Download | -------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK-EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEF----FGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.20 | 0.20 | 0.70 | 1.69 | Download | -----------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFMNLYTLY-TVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWEDGECYIQFFS---NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP---------C----IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------------------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.20 | 0.17 | 0.74 | 4.39 | Download | -------------------------------PCQKINVQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK-----KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN----CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.19 | 0.17 | 0.76 | 3.23 | Download | ------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE-KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQ----EFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.22 | 0.19 | 0.74 | 3.52 | Download | --------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMAGSIDCTLTFSHWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKERRITRMVLVVVAVFIVCWTPIHIYVIIKAL----------ITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
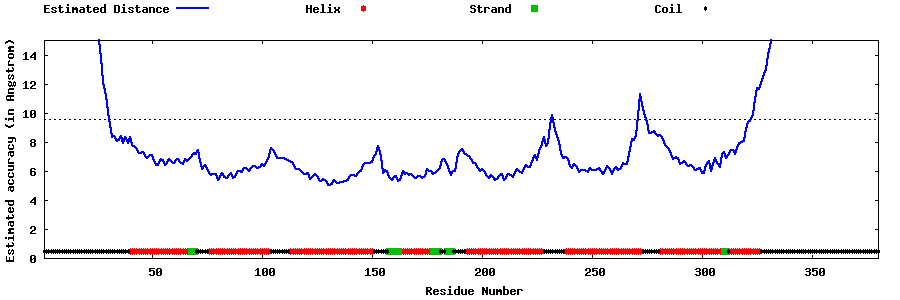
| Generated 3D models | Estimated local accuracy of models | ||
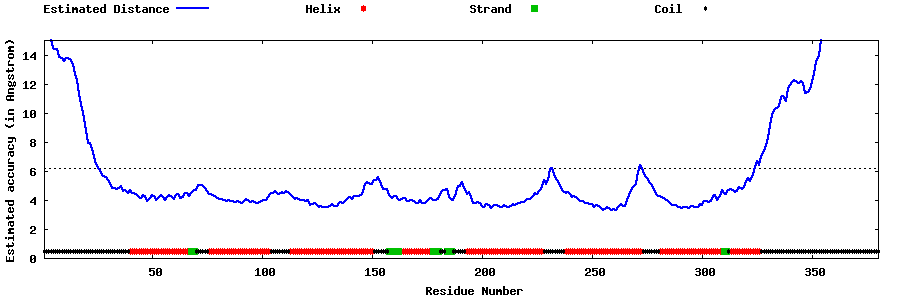 | |||
|
|
|||
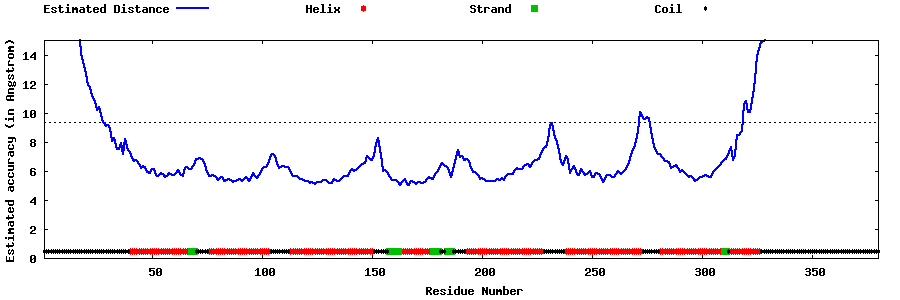 | |||
|
| |||
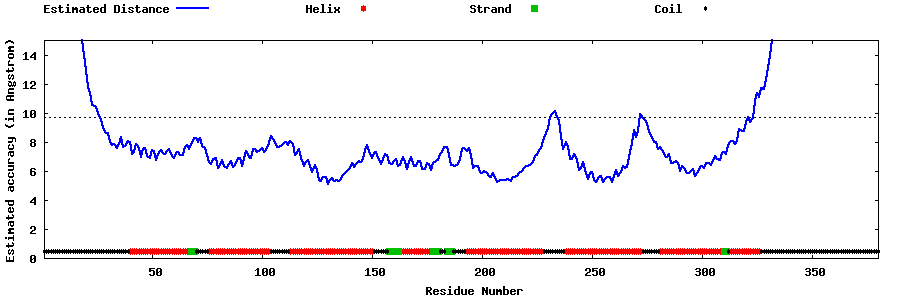 | |||
|
| |||
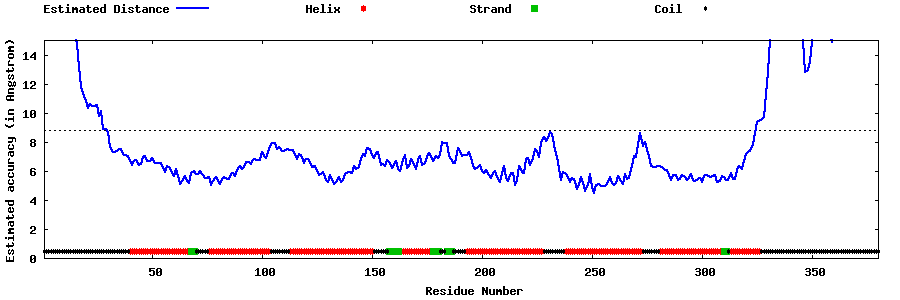 | |||
|
| |||
 | |||
|
| |||