

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MALTPESPSSFPGLAATGSSVPEPPGGPNATLNSSWASPTEPSSLEDLVATGTIGTLLSAMGVVGVVGNAYTLVVTCRSLRAVASMYVYVVNLALADLLYLLSIPFIVATYVTKEWHFGDVGCRVLFGLDFLTMHASIFTLTVMSSERYAAVLRPLDTVQRPKGYRKLLALGTWLLALLLTLPVMLAMRLVRRGPKSLCLPAWGPRAHRAYLTLLFATSIAGPGLLIGLLYARLARAYRRSQRASFKRARRPGARALRLVLGIVLLFWACFLPFWLWQLLAQYHQAPLAPRTARIVNYLTTCLTYGNSCANPFLYTLLTRNYRDHLRGRVRGPGSGGGRGPVPSLQPRARFQRCSGRSLSSCSPQPTDSLVLAPAAPARPAPEGPRAPA | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 98899999878777777788878889987687777778777543104579998999999999999888998667541388878387999999999999999968999999973898897688989999999999999999999999847863222764546756788540799999999998999982589709927982789904899999999999999999999999999999995457852012212031388999999999999989999999999996421316899999999999999999798999998498999999998454256877787887686566457678867788989888666688899999898999998 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MALTPESPSSFPGLAATGSSVPEPPGGPNATLNSSWASPTEPSSLEDLVATGTIGTLLSAMGVVGVVGNAYTLVVTCRSLRAVASMYVYVVNLALADLLYLLSIPFIVATYVTKEWHFGDVGCRVLFGLDFLTMHASIFTLTVMSSERYAAVLRPLDTVQRPKGYRKLLALGTWLLALLLTLPVMLAMRLVRRGPKSLCLPAWGPRAHRAYLTLLFATSIAGPGLLIGLLYARLARAYRRSQRASFKRARRPGARALRLVLGIVLLFWACFLPFWLWQLLAQYHQAPLAPRTARIVNYLTTCLTYGNSCANPFLYTLLTRNYRDHLRGRVRGPGSGGGRGPVPSLQPRARFQRCSGRSLSSCSPQPTDSLVLAPAAPARPAPEGPRAPA | |
| 76342432222330333233243333331222232333333423233100000002201200220131112000000103322100000000002021000000000000002430100200110010013210100000000000000000000020231232100000000110000000000000202537422101010256121000001023113310200020000000101313444444443421000000000000000012110000001001323223200100100000000100010000000004300410140021224444454443344444344344334444444334334134444444346346338 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MALTPESPSSFPGLAATGSSVPEPPGGPNATLNSSWASPTEPSSLEDLVATGTIGTLLSAMGVVGVVGNAYTLVVTCRSLRAVASMYVYVVNLALADLLYLLSIPFIVATYVTKEWHFGDVGCRVLFGLDFLTMHASIFTLTVMSSERYAAVLRPLDTVQRPKGYRKLLALGTWLLALLLTLPVMLAMRLVRRGPKSLCLPAWGPRAHRAYLTLLFATSIAGPGLLIGLLYARLARAYRRSQRASFKRARRPGARALRLVLGIVLLFWACFLPFWLWQLLAQYHQAPLAPRTARIVNYLTTCLTYGNSCANPFLYTLLTRNYRDHLRGRVRGPGSGGGRGPVPSLQPRARFQRCSGRSLSSCSPQPTDSLVLAPAAPARPAPEGPRAPA | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.28 | 0.24 | 0.76 | 3.23 | Download | --------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------ | |||||||||||||||||||
| 2 | 4n6hA | 0.28 | 0.25 | 0.76 | 3.85 | Download | -------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP-------------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.28 | 0.24 | 0.76 | 3.70 | Download | --------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------ | |||||||||||||||||||
| 4 | 2ziy | 0.18 | 0.20 | 0.88 | 1.50 | Download | ----------------------------DLRDNETWWYNPSIFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTEGVLCNCSFDYISRDTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV---TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETED------D---KDAETEIPAG-ESSDAAPSADAAQMKE------ | |||||||||||||||||||
| 5 | 4djh | 0.29 | 0.25 | 0.72 | 1.16 | Download | ----------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDDYLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.28 | 0.24 | 0.76 | 3.37 | Download | ----------------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------ | |||||||||||||||||||
| 7 | 4djh | 0.28 | 0.25 | 0.71 | 1.69 | Download | -------------------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF---------------------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.28 | 0.24 | 0.77 | 4.77 | Download | ------------------------------------GSPG-ARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKLDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------ | |||||||||||||||||||
| 9 | 4n6hA | 0.28 | 0.24 | 0.76 | 3.27 | Download | --------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------ | |||||||||||||||||||
| 10 | 5c1mA | 0.27 | 0.23 | 0.75 | 3.81 | Download | -----------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF--------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
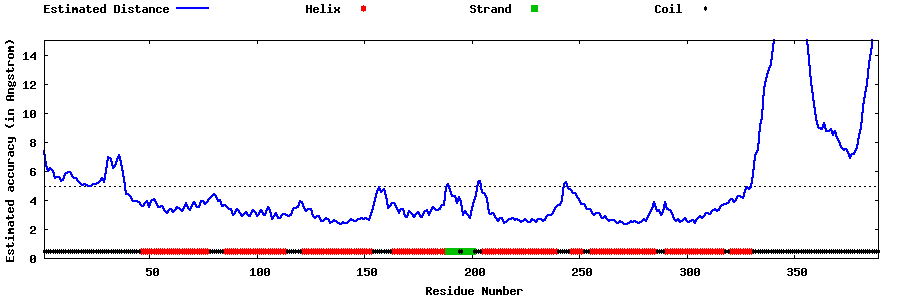 | |||
|
|
|||
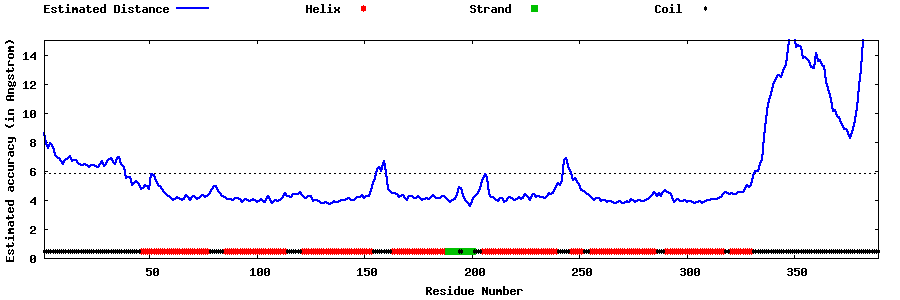 | |||
|
| |||
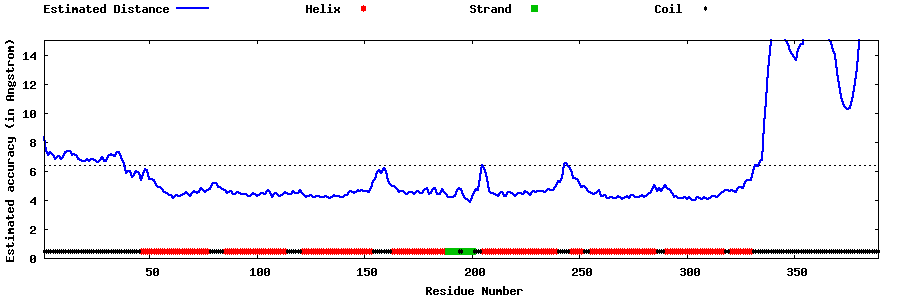 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||