

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MTALSSENCSFQYQLRQTNQPLDVNYLLFLIILGKILLNILTLGMRRKNTCQNFMEYFCISLAFVDLLLLVNISIILYFRDFVLLSIRFTKYHICLFTQIISFTYGFLHYPVFLTACIDYCLNFSKTTKLSFKCQKLFYFFTVILIWISVLAYVLGDPAIYQSLKAQNAYSRHCPFYVSIQSYWLSFFMVMILFVAFITCWEEVTTLVQAIRITSYMNETILYFPFSSHSSYTVRSKKIFLSKLIVCFLSTWLPFVLLQVIIVLLKVQIPAYIEMNIPWLYFVNSFLIATVYWFNCHKLNLKDIGLPLDPFVNWKCCFIPLTIPNLEQIEKPISIMIC | |
| CCCCCCCCCCHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCSSHHHHHHHHHHHHCCCCCCSSSSSCHHHHSSHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCSSSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSSSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHSSCCCHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCSSSSSCCCCCCCCCCCCCCSSSSC | |
| 98556677651003345799987412125999879999885312113675056788899999999999999999999987663045541221028999999997324577522474127640177743687567889999999999999999982687555566778876777875885047788899999999999988999999999999987347438996237898777754877899999999512178999999999973875031233742899988999998874026766622355777876661587642656886557898247649 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MTALSSENCSFQYQLRQTNQPLDVNYLLFLIILGKILLNILTLGMRRKNTCQNFMEYFCISLAFVDLLLLVNISIILYFRDFVLLSIRFTKYHICLFTQIISFTYGFLHYPVFLTACIDYCLNFSKTTKLSFKCQKLFYFFTVILIWISVLAYVLGDPAIYQSLKAQNAYSRHCPFYVSIQSYWLSFFMVMILFVAFITCWEEVTTLVQAIRITSYMNETILYFPFSSHSSYTVRSKKIFLSKLIVCFLSTWLPFVLLQVIIVLLKVQIPAYIEMNIPWLYFVNSFLIATVYWFNCHKLNLKDIGLPLDPFVNWKCCFIPLTIPNLEQIEKPISIMIC | |
| 74424375130323244344213010000000323131000000023461343000000000031101110000001101100000021031000000100010223021100000001100101312313430210000000321210000000212212431637622323010101111210000002101100100132001003002021134300000000234433143331001000100212330223201000003030101010200000000000000000030340403633031201000200000030742643732110000 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCSSHHHHHHHHHHHHCCCCCCSSSSSCHHHHSSHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCSSSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSSSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHSSCCCHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCSSSSSCCCCCCCCCCCCCCSSSSC MTALSSENCSFQYQLRQTNQPLDVNYLLFLIILGKILLNILTLGMRRKNTCQNFMEYFCISLAFVDLLLLVNISIILYFRDFVLLSIRFTKYHICLFTQIISFTYGFLHYPVFLTACIDYCLNFSKTTKLSFKCQKLFYFFTVILIWISVLAYVLGDPAIYQSLKAQNAYSRHCPFYVSIQSYWLSFFMVMILFVAFITCWEEVTTLVQAIRITSYMNETILYFPFSSHSSYTVRSKKIFLSKLIVCFLSTWLPFVLLQVIIVLLKVQIPAYIEMNIPWLYFVNSFLIATVYWFNCHKLNLKDIGLPLDPFVNWKCCFIPLTIPNLEQIEKPISIMIC | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.15 | 0.19 | 0.85 | 1.41 | Download | -ENFMDIECFMVLNSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRC--RPSYHFIGSLAVADLLGSVIFVYSFIDFHVF----RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL-PLLGWNCEKLQSVC--SDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD----------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM----F--------------------------- | |||||||||||||||||||
| 2 | 4z34A | 0.13 | 0.20 | 0.86 | 2.43 | Download | -QCFYNESIAFFYNRSGKHLATEWNTVCIFIMLANLLVMVAIY---VNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVAN---LLAIAIERHITVFRM-QLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLY-SDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLAAEQLKTTRNAYIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMS-------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.13 | 0.20 | 0.88 | 2.53 | Download | ----SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTL---PFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMA--VTRPRDGAVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV---RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.13 | 0.20 | 0.97 | 1.53 | Download | ---DLRDNETWWYNPDQDAVYYSLGIFIGICGIIGCGGNGIVIYLKTKSL-QTPANMFIINLAFSDFTFSLVNFPLMTISC--FLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAI-GP-IFGWGAYTLGVLCNCSFDYDTRSNILCMLGFFGPILIIFFCYFNIVMVSNHEKEMAAMAKRLNAKELRKAQAGANAEMRKISIVIVSQFLLSWSPYAVVALLAQFGPLWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVL---TCCQFDDKETDAETEIPASSDAAP | |||||||||||||||||||
| 5 | 5xsz | 0.12 | 0.19 | 0.86 | 1.14 | Download | ----------------DNFKYPLYSMVFSIVFMVGLITNVAAMYIFCSLKLRNETTTYMMNLVVSDLLFVLTLPLRVF--YFV-QQNWPFGSLLCKLSVSLFYTNMYGSILFLTCISVDRFLAIVYPFRSRGLRTKRNAKIVCAAVWVLVLSGSL--PTGFMNTNKLENNSISCFEWKSHSKVVIFIETVLIPLMLNVVCSAMVLQTLRRPMLRIDNGLRL-KIYKNTEWDAYLNKKKRMIIVHLFIFCFCFIPYNVNLVFYSLVRTSVVRTIYPIALCIAVSNCCFDPIVYYFTSETI--QNSAS--SEDLYFQ----------------------- | |||||||||||||||||||
| 6 | 5tjvA | 0.14 | 0.19 | 0.85 | 2.21 | Download | ---MDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCR--PSYHFIGSLAVADLLGSVIFVYSFIDFHVF---HRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPH-IDETYLMFWIGVTSVLLLFIVYAYMYILWKAG------------KRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------------------- | |||||||||||||||||||
| 7 | 4z34A | 0.14 | 0.18 | 0.72 | 1.46 | Download | --------------------------LGITVCIFIMLANLLVMVAYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTG--PNTR-RLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFR-MQLHTRMSNRRVVVVIVVIWTMAIVMGAI-PSVGWN---CICDIENCSNMAPSDSYLVFWAIFNLVTF--------VVMVVLYAHY--------------LRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCP-QCDLAYEKFFLLLAEFNSAMNPIIYSYRDKEM--------------------------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.19 | 0.19 | 0.76 | 1.54 | Download | ------------------NSDLDVNIYLALFVVG-TVGNSVTLFTLARKKSQSTVDYYLGSLALSDLLILLLMPVELYNFIWVHHPWAF-GDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHKTLMSSRTKKF-----ISAIWLASALLAI--------PLSGDGTHPGGLVCTPIATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQ--------PGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQYHYFYMLTNALVYVSAAINPILYNLVSSTL--------------------------------------- | |||||||||||||||||||
| 9 | 3uonA | 0.14 | 0.18 | 0.79 | 1.61 | Download | ---------TFEV----VFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQ---TVNNYFLFSLACADLIIGVFSMNLYTLYTVI--GYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPA-QFIVGVRTVEDGECYIQFFSNAVTFGTAIAAFYLPVIIMTVLYWHISRASK---------------------SRREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------ | |||||||||||||||||||
| 10 | 3zpqA | 0.16 | 0.19 | 0.83 | 1.04 | Download | ----GAELLSQQWEAGMS---LLMALVVLLIVAGNVLVIAAIGSTQRLQTLT---NLFITSLACADLVVGL--LVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQQDPGCCDFVTNRAYAIASSIIYIPLLIMIFVALRVYREAKEQSRVMLMRE---------------HKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCRS-PDFRKAFKRLLA------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
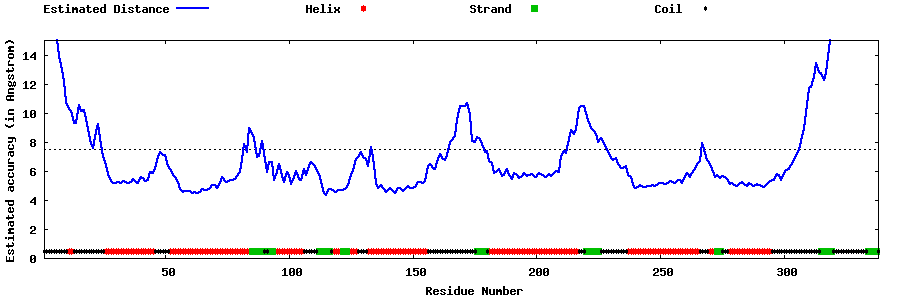
| Generated 3D models | Estimated local accuracy of models | ||
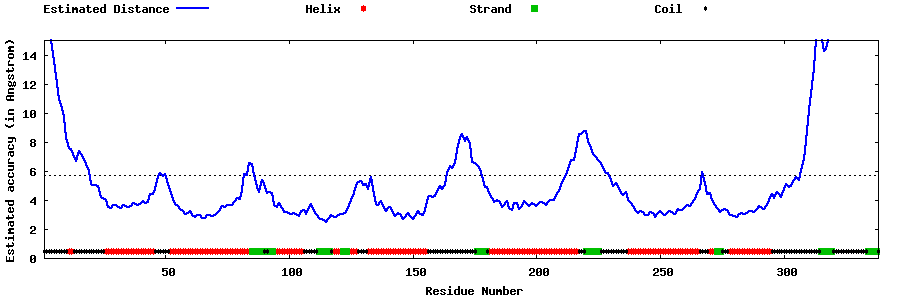 | |||
|
|
|||
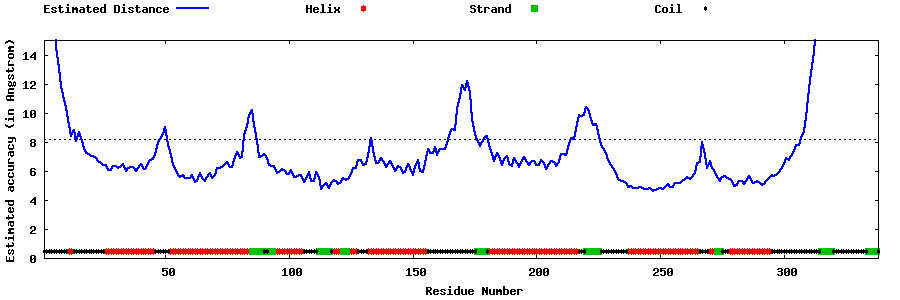 | |||
|
| |||
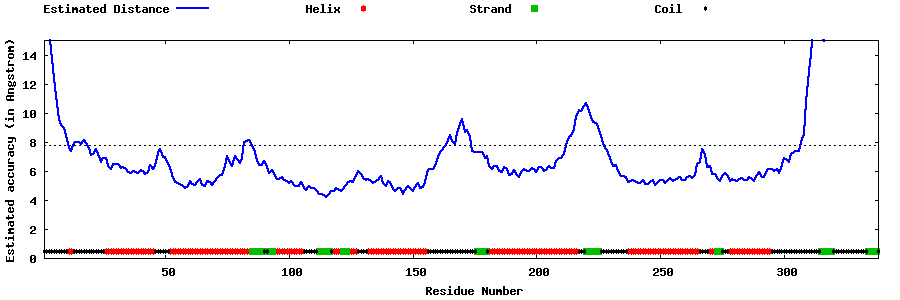 | |||
|
| |||
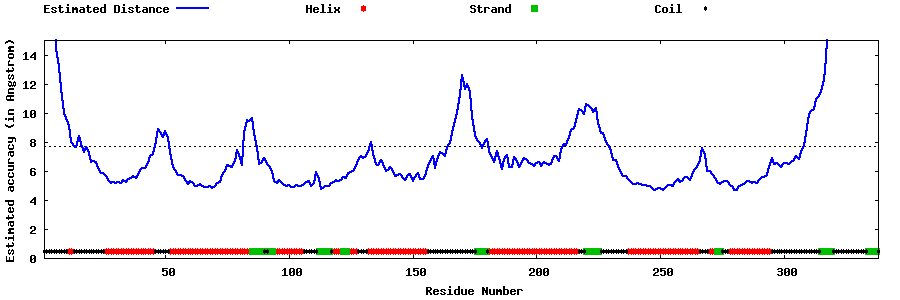 | |||
|
| |||
 | |||
|
| |||