

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MVNLTSMSGFLLMGFSDERKLQILHALVFLVTYLLALTGNLLIITIITVDRRLHSPMYYFLKHLSLLDLCFISVTVPQSIANSLMGNGYISLVQCILQVFFFIALASSEVAILTVMSYDRYAAICQPLHYETIMDPRACRHAVIAVWIAGGLSGLMHAAINFSIPLCGKRVIHQFFCDVPQMLKLACSYEFINEIALAAFTTSAAFICLISIVLSYIRIFSTVLRIPSAEGRTKVFSTCLPHLFVATFFLSAAGFEFLRLPSDSSSTVDLVFSVFYTVIPPTLNPVIYSLRNDSMKAALRKMLSKEELPQRKMCLKAMFKL | |
| CCCCCCHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHCCHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHCCCCHHHHHHHHHHCC | |
| 998862277631158998507999999999999999998999999996188756748999877999856255025899999871599768789999999999999999999999998750776175554772337869999999999999999999999981527999593789666868888771655349899999999999999899899999999999720866267664203708699999999854650670799989887780899852055524345532256199999999998626241333288887149 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MVNLTSMSGFLLMGFSDERKLQILHALVFLVTYLLALTGNLLIITIITVDRRLHSPMYYFLKHLSLLDLCFISVTVPQSIANSLMGNGYISLVQCILQVFFFIALASSEVAILTVMSYDRYAAICQPLHYETIMDPRACRHAVIAVWIAGGLSGLMHAAINFSIPLCGKRVIHQFFCDVPQMLKLACSYEFINEIALAAFTTSAAFICLISIVLSYIRIFSTVLRIPSAEGRTKVFSTCLPHLFVATFFLSAAGFEFLRLPSDSSSTVDLVFSVFYTVIPPTLNPVIYSLRNDSMKAALRKMLSKEELPQRKMCLKAMFKL | |
| 772403101000000043340011002303321331333332002001004300000010030000000101000002000010154320001001101100000023000000100100000002002010301330000000000210231021000000002004623020000012200300012230101101221233133122203302320010001031663231000001102000101320021000103163344241000011023313300300002042014002200333303443010300043 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHCCHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHCCCCHHHHHHHHHHCC MVNLTSMSGFLLMGFSDERKLQILHALVFLVTYLLALTGNLLIITIITVDRRLHSPMYYFLKHLSLLDLCFISVTVPQSIANSLMGNGYISLVQCILQVFFFIALASSEVAILTVMSYDRYAAICQPLHYETIMDPRACRHAVIAVWIAGGLSGLMHAAINFSIPLCGKRVIHQFFCDVPQMLKLACSYEFINEIALAAFTTSAAFICLISIVLSYIRIFSTVLRIPSAEGRTKVFSTCLPHLFVATFFLSAAGFEFLRLPSDSSSTVDLVFSVFYTVIPPTLNPVIYSLRNDSMKAALRKMLSKEELPQRKMCLKAMFKL | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.22 | 0.87 | 3.55 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINC-FTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ----------- | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.22 | 0.85 | 2.15 | Download | -ENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM------------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.19 | 0.22 | 0.83 | 2.07 | Download | ----------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH--------------------ILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.17 | 0.20 | 0.86 | 1.54 | Download | ---------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIK--GIE----TNPNNITCV----------LTKEFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLNEQRASKVLGIVFFLFLLMWCPFFITNITVLCDSNQQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.23 | 0.88 | 1.23 | Download | KTRNAYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNV-FF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------------------ | |||||||||||||||||||
| 6 | 3emlA | 0.19 | 0.22 | 0.86 | 3.76 | Download | -------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ----------- | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.23 | 0.82 | 1.71 | Download | -------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWR-QASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLRERKATKTLG--IILGAFIVCWLPFFIISLVMPIHL-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.19 | 0.19 | 0.79 | 2.75 | Download | -----------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAI--------MGSAQVEIPTPQDYWGPVFAICIFLFSF-----------------IVPVLVISVCYSLMIRRLRGVRLLSGSVAVFVGCWTPVQVFVLAQGLG-----VQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR--------------------- | |||||||||||||||||||
| 9 | 3emlA | 0.19 | 0.22 | 0.86 | 4.60 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIV-GLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ----------- | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.21 | 0.93 | 5.72 | Download | ---------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLPFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLPFKAAAAENLYF- | |||||||||||||||||||
| ||||||||||||||||||||||||||
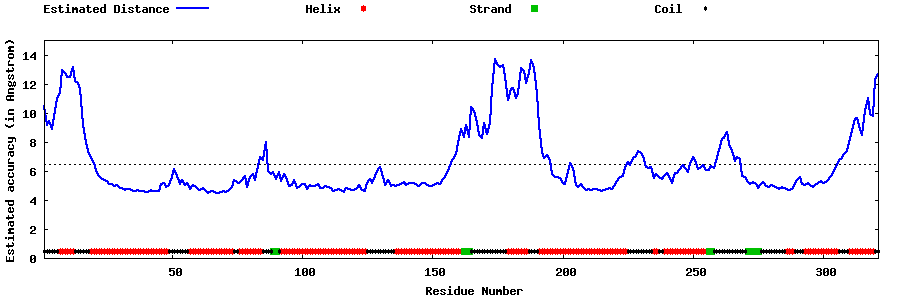
| Generated 3D models | Estimated local accuracy of models | ||
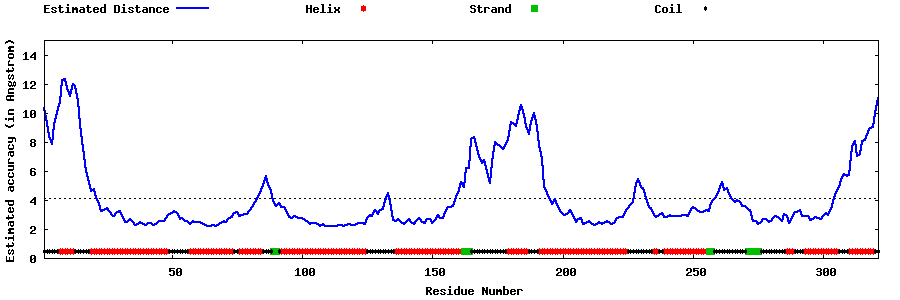 | |||
|
|
|||
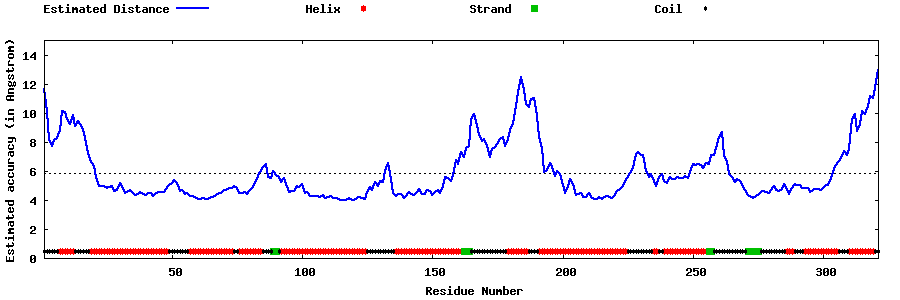 | |||
|
| |||
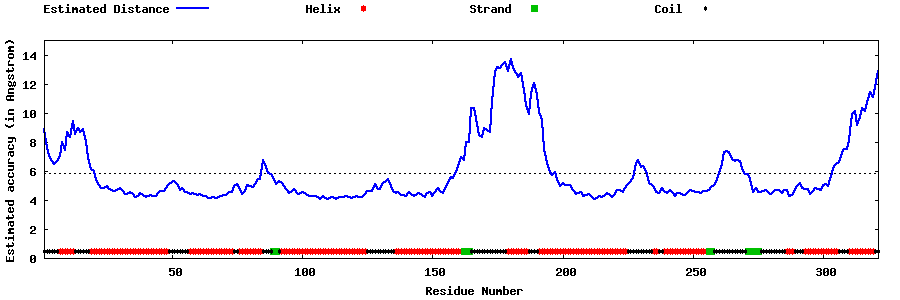 | |||
|
| |||
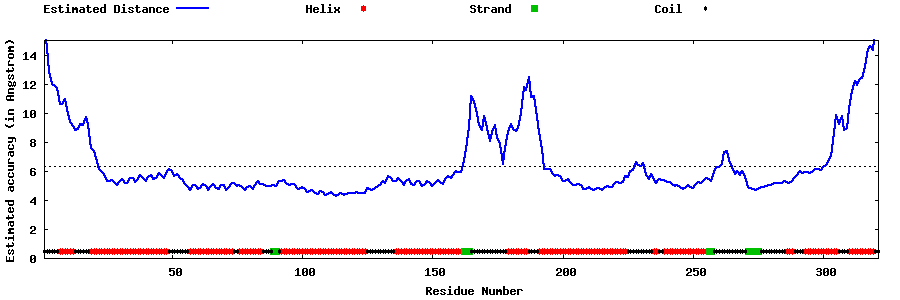 | |||
|
| |||
 | |||
|
| |||