

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGNDSVSYEYGDYSDLSDRPVDCLDGACLAIDPLRVAPLPLYAAIFLVGVPGNAMVAWVAGKVARRRVGATWLLHLAVADLLCCLSLPILAVPIARGGHWPYGAVGCRALPSIILLTMYASVLLLAALSADLCFLALGPAWWSTVQRACGVQVACGAAWTLALLLTVPSAIYRRLHQEHFPARLQCVVDYGGSSSTENAVTAIRFLFGFLGPLVAVASCHSALLCWAARRCRPLGTAIVVGFFVCWAPYHLLGLVLTVAAPNSALLARALRAEPLIVGLALAHSCLNPMLFLYFGRAQLRRSLPAACHWALRESQGQDESVDSKKSTSHDLVSEMEV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSSHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9998888877776567788877666553455504646999999999998855786020011146666899999999999999999839999999758988797707989999999999999999999999836752111402127662510000799999999998499997520560789848997159960689999999999999999999999999999999987786863010078999978999999999983068637899999999999999999848589898208788999999982721367877787888888887787756419 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGNDSVSYEYGDYSDLSDRPVDCLDGACLAIDPLRVAPLPLYAAIFLVGVPGNAMVAWVAGKVARRRVGATWLLHLAVADLLCCLSLPILAVPIARGGHWPYGAVGCRALPSIILLTMYASVLLLAALSADLCFLALGPAWWSTVQRACGVQVACGAAWTLALLLTVPSAIYRRLHQEHFPARLQCVVDYGGSSSTENAVTAIRFLFGFLGPLVAVASCHSALLCWAARRCRPLGTAIVVGFFVCWAPYHLLGLVLTVAAPNSALLARALRAEPLIVGLALAHSCLNPMLFLYFGRAQLRRSLPAACHWALRESQGQDESVDSKKSTSHDLVSEMEV | |
| 7646434131322332333423246442342300100002212200230232122000000113421000000000010300000000010010034442100100110010013211110000000001000000000020232232110100000011000000100000020343645220101030444520210010211120233112000300000022134201000000000201021311100001002334142231021010100000120002101000000362024002300330044444464444454444454444447 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSSHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MGNDSVSYEYGDYSDLSDRPVDCLDGACLAIDPLRVAPLPLYAAIFLVGVPGNAMVAWVAGKVARRRVGATWLLHLAVADLLCCLSLPILAVPIARGGHWPYGAVGCRALPSIILLTMYASVLLLAALSADLCFLALGPAWWSTVQRACGVQVACGAAWTLALLLTVPSAIYRRLHQEHFPARLQCVVDYGGSSSTENAVTAIRFLFGFLGPLVAVASCHSALLCWAARRCRPLGTAIVVGFFVCWAPYHLLGLVLTVAAPNSALLARALRAEPLIVGLALAHSCLNPMLFLYFGRAQLRRSLPAACHWALRESQGQDESVDSKKSTSHDLVSEMEV | |||||||||||||||||||||||||
| 1 | 5o9hA | 0.38 | 0.36 | 0.85 | 3.33 | Download | ----------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVLCGVDHD--KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAG-QGFQKSLPELLREVLTEESVVR------------------- | |||||||||||||||||||
| 2 | 5vblA | 0.25 | 0.26 | 0.84 | 3.94 | Download | -----------------------------DWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSSRRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLRLRVSGAVATAVLWVLAALLAMPVMVLRTTGDLENTNKVQCYMDYSSEWAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIRRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLPCDFDLFLMNIFPYCTCISYVNSCLNPFLYAFFDPR-FRQACTSMLLMGQSR------------------------ | |||||||||||||||||||
| 3 | 5o9hA | 0.38 | 0.36 | 0.85 | 3.78 | Download | ----------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVLCGV--DHDKRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGF-QKSLPELLREVLTEESVVR------------------- | |||||||||||||||||||
| 4 | 4djh | 0.21 | 0.22 | 0.80 | 1.52 | Download | ------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRRITRLVVVAVFVVCWTPIHIFILVEALGSA-------ALSSYYFCIALGYTNSSLNPILYAFLDE-NFKRCFRDFCFP---------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.21 | 0.22 | 0.80 | 1.16 | Download | ------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA--A-----LSSYYFCIALGYTNSSLNPILYAFLDE-NFKRCFRDFCFP---------------------------- | |||||||||||||||||||
| 6 | 5o9hA | 0.38 | 0.36 | 0.85 | 3.33 | Download | -----------------------------TLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVLCGVDHD--KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAG-QGFQKSLPELLREVLTEESVVR------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.19 | 0.22 | 0.81 | 1.66 | Download | ------------------------------PDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEG-VLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEW----VTPYAAQLPVMFAKASAIHNPMIYSVSHP-KFREAISQTFPW---------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.23 | 0.23 | 0.82 | 4.28 | Download | ------------------------------GSPLAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPSPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-PLVVAALHLCIALGYANSSLNPVLYAFLD-ENFKRCFRQLCRKPCG------------------------- | |||||||||||||||||||
| 9 | 5o9hA | 0.38 | 0.36 | 0.85 | 3.42 | Download | ----------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVLCGVDHD--KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVA-GQGFQKSLPELLREVLTEESVVR------------------- | |||||||||||||||||||
| 10 | 5o9hA | 0.38 | 0.36 | 0.85 | 4.18 | Download | ----------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVLCGVDHD--KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQG-FQKSLPELLREVLTEESVVR------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
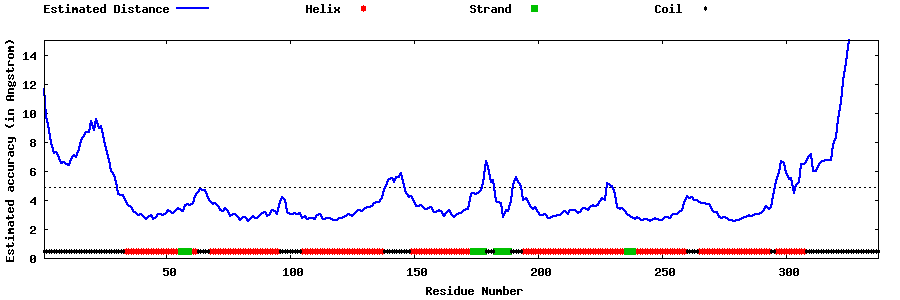 | |||
|
|
|||
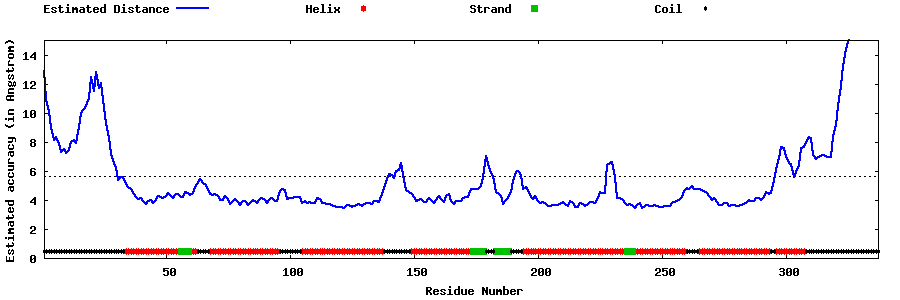 | |||
|
| |||
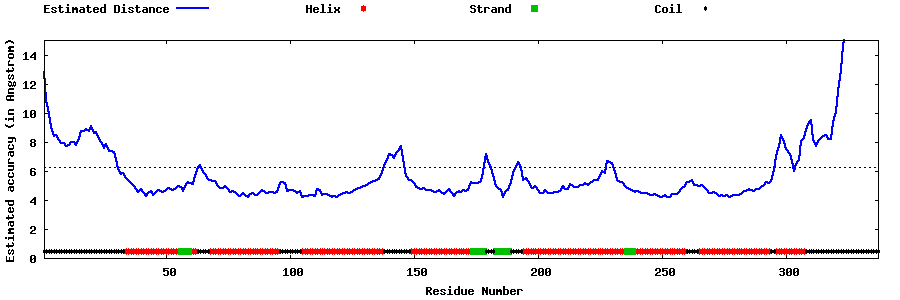 | |||
|
| |||
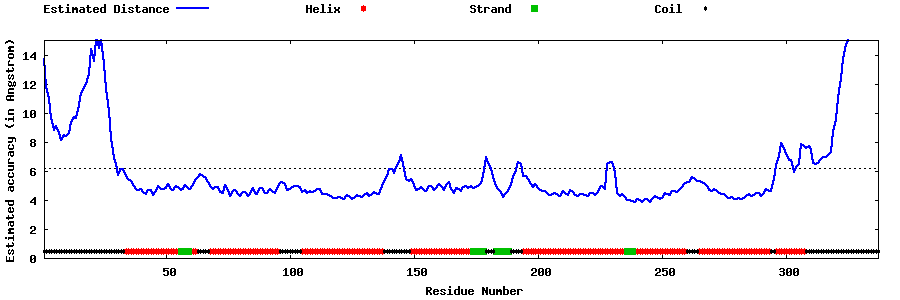 | |||
|
| |||
 | |||
|
| |||